化工学报 ›› 2019, Vol. 70 ›› Issue (11): 4337-4345.DOI: 10.11949/0438-1157.20190278
郑禾1( ),杨盛江2,郑永超1,崔燕1,郭旋1,钟近艺1(
),杨盛江2,郑永超1,崔燕1,郭旋1,钟近艺1( ),周健2(
),周健2( )
)
收稿日期:2019-03-22
修回日期:2019-07-10
出版日期:2019-11-05
发布日期:2019-11-05
通讯作者:
钟近艺,周健
作者简介:郑禾(1984—),男,博士研究生,助理研究员,基金资助:
He ZHENG1( ),Shengjiang YANG2,Yongchao ZHENG1,Yan CUI1,Xuan GUO1,Jinyi ZHONG1(
),Shengjiang YANG2,Yongchao ZHENG1,Yan CUI1,Xuan GUO1,Jinyi ZHONG1( ),Jian ZHOU2(
),Jian ZHOU2( )
)
Received:2019-03-22
Revised:2019-07-10
Online:2019-11-05
Published:2019-11-05
Contact:
Jinyi ZHONG,Jian ZHOU
摘要:
DhaA能够有效降解化学毒剂芥子气,而环境耐受性差影响了其在军事洗消中的应用。虽然已有研究表明定向进化、化学修饰、固定化有利于提高DhaA在尿素、二甲基亚砜(DMSO)溶液中的稳定性,但DhaA在尿素、DMSO下的变性过程尚不清晰。利用分子动力学(MD)模拟方法研究了DhaA在尿素和DMSO两种体系中的变性过程,结果表明尿素分子通过取代水分子与DhaA形成氢键的方式诱导其变性,并且能够与催化位点形成氢键,造成DhaA底物进出口通道长度增加、通道曲率增大、瓶颈尺寸减小;DMSO分子通过范德华作用进入DhaA疏水空腔,从而诱导DhaA变性,使得DhaA通道长度缩短、瓶颈尺寸增大,造成DhaA发生构象变化。该研究结果揭示了DhaA在两种体系中变性过程的区别,能够为DhaA的进一步稳定化提供理论指导。
中图分类号:
郑禾, 杨盛江, 郑永超, 崔燕, 郭旋, 钟近艺, 周健. 尿素和二甲基亚砜诱导DhaA变性的分子动力学模拟[J]. 化工学报, 2019, 70(11): 4337-4345.
He ZHENG, Shengjiang YANG, Yongchao ZHENG, Yan CUI, Xuan GUO, Jinyi ZHONG, Jian ZHOU. Molecular dynamics simulation of denaturation of DhaA induced by urea and dimethyl sulfoxide[J]. CIESC Journal, 2019, 70(11): 4337-4345.
| DhaA模拟体系 | 通道长度/? | 通道曲率 | 瓶颈尺寸/? | 通过成本 |
|---|---|---|---|---|
| 纯水 | 12.55 | 1.25 | 1.16 | 0.45 |
| 尿素 | 22.48 | 1.45 | 1.10 | 0.54 |
| DMSO | 10.27 | 1.27 | 1.26 | 0.19 |
表1 DhaA在不同变性条件下的主通道结构参数
Table 1 Main tunnel parameters of DhaA in different simulation systems
| DhaA模拟体系 | 通道长度/? | 通道曲率 | 瓶颈尺寸/? | 通过成本 |
|---|---|---|---|---|
| 纯水 | 12.55 | 1.25 | 1.16 | 0.45 |
| 尿素 | 22.48 | 1.45 | 1.10 | 0.54 |
| DMSO | 10.27 | 1.27 | 1.26 | 0.19 |
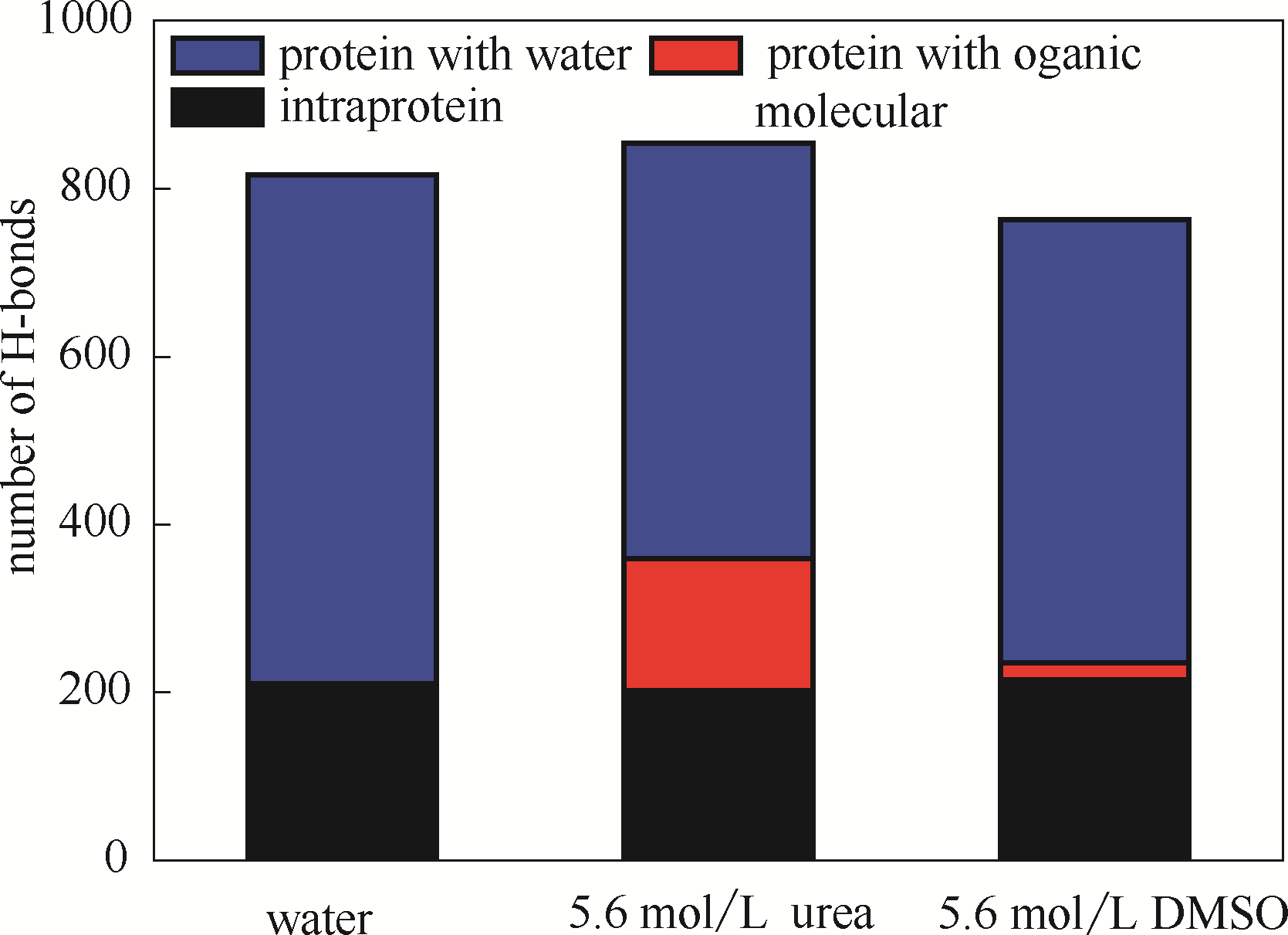
图5 DhaA在不同模拟体系中的氢键数(蓝色表示DhaA与水分子氢键,红色表示DhaA与有机分子氢键,黑色表示DhaA分子内氢键)
Fig.5 Numbers of H-bonds in DhaA in simulation systems(blue bar represents H-bond between DhaA and water molecules, red bar represents H-bond between DhaA and organic molecules, black bar represents intramolecular H-bond of DhaA molecules)
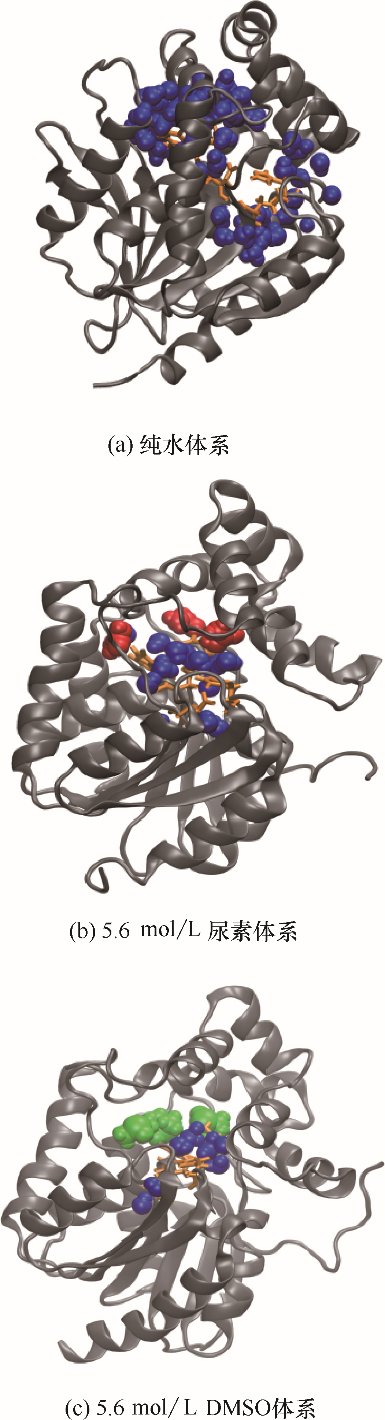
图6 DhaA在不同模拟体系中的催化位点3.5 ?内溶剂分子分布(蓝色珠子代表水分子,红色珠子代表尿素分子,绿色珠子代表DMSO分子)
Fig.6 Snapshots of solution molecules distribution within 3.5 ? around catalytic sites in DhaA in different simulation systems(blue beads represent water molecules, red beads represent urea molecules, green beads represent DMSO molecules)

图8 DhaA在模拟体系中催化位点的结构变化(蓝色为纯水体系中的DhaA,红色为尿素体系中的DhaA,橙色为DMSO体系中的DhaA)
Fig.8 Structural change of catalytic sites of DhaA in different simulation systems(blue part represents DhaA in water, red part represents DhaA in urea solution, orange part represents DhaA in DMSO solution)
| 1 | KoudelakovaT, BidmanovaS, DvorakP, et al. Haloalkane dehalogenases: biotechnological applications[J]. Biotechnol. J., 2013, 8: 32-45. |
| 2 | NagataY, OhtsuboY, TsudaM. Properties and biotechnological applications of natural and engineered haloalkane dehalogenases[J]. Appl. Microbiol. Biotechnol., 2015, 99: 9865-9881. |
| 3 | HarveyP S. Enzymatic degradation of HD[R]. USA: Edgewood Chemical Biological Center, 2002. |
| 4 | BidmanovaS, SteinerM S, StepanM, et al. Enzyme-based test strips for visual or photographic detection and quantitation of gaseous sulfur mustard[J]. J. Anal. Chem., 2016, 88: 6044-6049. |
| 5 | 郭楠, 董亮, 刘景全, 等. 烷基卤去卤化酶对芥子气的催化水解[J]. 环境化学, 2015, 34: 1363-1370. |
| GuoN, DongL, LiuJ Q, et al. Catalytic hydrolysis of sulfur mustard by haloalkane dehalogenases[J]. Environ. Chem., 2015, 34: 1363-1370. | |
| 6 | 赵渊中, 钟近艺, 郭楠, 等. 多点突变提高DhaA对芥子气的活性和热稳定性[J]. 应用与环境生物学报, 2017, 23: 714-718. |
| ZhaoY Z, ZhongJ Y, GuoN, et al. Improvement in the thermostability and activity of DhaA against sulfur mustard by multipoint mutagenesis[J]. Chin. J. Appl. Environ. Biol., 2017, 23: 714-718. | |
| 7 | StepankovaV, DamborskyJ, ChaloupkovaR. Organic co-solvents affect activity, stability and enantioselectivity of haloalkane dehalogenases[J]. Biotechnol. J., 2013, 8: 719-729. |
| 8 | LiskovaV, BednarD, PrudnikovaT, et al. Balancing the stability-activity trade-off by fine-tuning dehalogenase access tunnels[J]. ChemCatChem, 2015, 7: 648-659. |
| 9 | ZhaoY Z, YuW L, ZhengH, et al. PEGylation with the thiosuccinimido butylamine linker significantly increases the stability of haloalkane dehalogenase DhaA[J]. J. Biotechnol., 2017, 254: 25-33. |
| 10 | ZhengH, ZhongJ Y, CuiY, et al. Mesoporous support designed for DhaA adsorption with improved stability[J]. J. Porous. Mat., 2019, 26(3): 829-837. |
| 11 | 郑禾, 钟近艺, 崔燕, 等. 荧光光谱法研究氨基改性介孔泡沫对DhaA的稳定化机理[J]. 光谱学与光谱分析, 2019, 39: 1776-1784. |
| ZhengH, ZhongJ Y, CuiY, et al. Stabilization mechanism of amino-mesocellular foam to DhaA by fluorescence spectroscopic method[J]. Spectrosc. Spect. Anal., 2019, 39: 1776-1784. | |
| 12 | DaggettV. Molecular dynamics simulations of the protein unfolding/folding reaction[J]. Acc. Chem. Res., 2002, 35: 422-429. |
| 13 | TretyakovaT, ShushanyanM, PartskhaladzeT, et al. Simplicity within the complexity: bilateral impact of DMSO on the functional and unfolding patterns of α-chymotrypsin[J]. Biophys. Chem., 2013, 175/176: 17-27. |
| 14 | KhanS H, PrakashA, PandeyP, et al. Protein folding: molecular dynamics simulations and in vitro studies for probing mechanism of urea- and guanidinium chloride induced unfolding of horse cytochrome-c[J]. Int. J. Biol. Macromol., 2019, 122: 695-704. |
| 15 | YamadaT, MitakuS, YamatoT. Characterization of mechanical unfolding intermediates of membrane proteins by coarse grained molecular dynamics simulation[J]. Chem. Phys. Lett., 2018, 691: 276-282. |
| 16 | CanchiD R, GarciaA E. Cosolvent effects on protein stability[J]. Annu. Rev. Phys. Chem., 2013, 64: 273-293. |
| 17 | 廖晨伊, 周健. β发卡多肽Trpzip4折叠的副本交换分子动力学模拟[J]. 化学学报, 2013, 71: 593-601. |
| LiaoC Y, ZhouJ. Replica exchange molecular dynamics simulations on the folding of Trpzip4 β-hairpin[J]. Acta Chim. Sinica, 2013, 71: 593-601. | |
| 18 | 曹了然, 张春煜, 张鼎林, 等. 分子动力学模拟技术在生物分子研究中的进展[J]. 物理化学学报, 2017, 33: 1354-1365. |
| CaoL R, ZhangC Y, ZhangD L, et al. Recent developments in using molecular dynamics simulation techniques to study biomolecules[J]. Acta Physico-Chimica Sinica, 2017, 33: 1354-1365. | |
| 19 | RoccatanoD, WongT S, SchwanebergU, et al. Structural and dynamic properties of cytochrome P450 BM-3 in pure water and in a dimethylsulfoxide/water mixture[J]. Biopolymers, 2005, 78: 259-267. |
| 20 | LiJ H, ChenY, YangJ, et al. Thermal and urea induced unfolding processes of glutathione S-transferase by molecular dynamics simulation[J]. Biopolymers, 2015, 103: 247-259. |
| 21 | KhanP, PrakashA, HaqueM A, et al. Structural basis of urea-induced unfolding: unraveling the folding pathway of hemochromatosis factor E[J]. Int. J. Biol. Macromol., 2016, 91: 1051-1061. |
| 22 | 沈洪辰, 丁吉勇, 李丽, 等. Y220C突变体影响p53C蛋白质构象转换的分子动力学模拟[J]. 物理化学学报, 2016, 32: 2620-2627. |
| ShenH C, DingJ Y, LiL, et al. Effect of Y220C mutant on the conformational transition of p53C probed by molecular dynamics simulation[J]. Acta Physico-Chimica Sinica, 2016, 32: 2620-2627. | |
| 23 | 卢滇楠, 闫明, 张敏莲, 等. 蛋白质-表面活性剂组装结构的分子模拟[J]. 化工学报, 2006, 57(8): 1949-1956. |
| LuD N, YanM, ZhangM L, et al. Molecular simulation of protein-surfactant assembly in aqueous solution[J]. Journal of Chemical Industry and Engineering(China), 2006, 57(8): 1949-1956. | |
| 24 | 杨程, 卢滇楠, 张敏莲, 等. 分子动力学模拟二硫键对胰岛素构象稳定性的影响[J]. 化工学报, 2010, 61(4): 929-934. |
| YangC, LuD N, ZhangM L, et al. Molecular dynamics simulation of impact of disulfide bridge on conformational stability of insulin[J]. CIESC Journal, 2010, 61(4): 929-934. | |
| 25 | 潘晓莉, 李代禧, 魏冬青. 胰岛素活性结构在水合离子液体中的稳定性[J]. 化工学报, 2017, 68(5): 2035-2041. |
| PanX L, LiD X, WeiD Q. Bioactive structural stability of insulin in hydrated ionic liquids[J]. CIESC Journal, 2017, 68(5): 2035-2041. | |
| 26 | HessB, KutznerC, van de SpoelD, et al. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation[J]. J. Chem. Theory Comput., 2008, 4: 435-447. |
| 27 | 赵渊中. 脱卤酶对芥子气的催化活性和稳定性研究[D]. 北京: 防化研究院, 2016. |
| ZhaoY Z. Study on the activity and stability of dehalogenase against sulfur mustard[D]. Beijing: Research Institute of Chemical Defense, 2016. | |
| 28 | JurcikA, BednarD, ByskaJ, et al. CAVER Analyst 2.0: analysis and visualization of channels and tunnels in protein structures and molecular dynamics trajectories[J]. Bioinfomatics, 2018, 34: 3586-3588. |
| 29 | AmreshP, GunjanD, NaveenK M, et al. Elucidation of stable intermediates in urea induced unfolding pathway of human carbonic anhydrase Ⅸ[J]. J. Biomol. Struct. Dtn., 2017, 36: 2391-2406. |
| [1] | 曾如宾, 沈中杰, 梁钦锋, 许建良, 代正华, 刘海峰. 基于分子动力学模拟的Fe2O3纳米颗粒烧结机制研究[J]. 化工学报, 2023, 74(8): 3353-3365. |
| [2] | 张永泉, 玄伟伟. 碱金属/(FeO+CaO+MgO)对硅酸盐灰熔渣结构和黏度的影响机理[J]. 化工学报, 2023, 74(4): 1764-1771. |
| [3] | 李泽严, 樊星, 李坚. 非热等离子体强化TiO2催化尿素分解副产物水解性能的研究[J]. 化工学报, 2021, 72(9): 4698-4707. |
| [4] | 刘万强,杨帆,袁华,张远达,易平贵,周虎. 醇类有机物热传导的分子动力学模拟及微观机理研究[J]. 化工学报, 2020, 71(11): 5159-5168. |
| [5] | 李兆宁, 赵彦杰, 汤玉鹏. 尿素水溶液凝固结晶附着力特性研究[J]. 化工学报, 2018, 69(S2): 232-239. |
| [6] | 张晋玮, 成洪业, 陈立芳, 漆志文. [BMIM]HSO4离子液体腐蚀性的实验与分子模拟[J]. 化工学报, 2018, 69(2): 808-814. |
| [7] | 李婷婷, 赵乐乐, 郑子良, 王振军, 张瑞平. 右旋布洛芬/尿素改性蒙脱土复合物的制备及体外释药性能[J]. 化工学报, 2017, 68(9): 3631-3637. |
| [8] | 徐上, 赵伶玲, 蔡庄立, 陈超. 二维氮化铝材料传热性能的模拟研究[J]. 化工学报, 2017, 68(9): 3321-3327. |
| [9] | 南怡伶, 孔宪, 李继鹏, 卢滇楠. 纳米狭缝中水流动非平衡分子动力学模拟[J]. 化工学报, 2017, 68(5): 1786-1793. |
| [10] | 冯志明, 李微微, 李雪, 赵阳, 谢晓峰, 柴春鹏, 罗运军. 不同羧酸根含量对双官能团聚降冰片烯质子交换膜性能影响的分子动力学模拟[J]. 化工学报, 2016, 67(S1): 253-259. |
| [11] | 单晨旭, 曹绪龙, 祝仰文, 刘坤, 曲广淼, 吕鹏飞, 薛春龙, 丁伟. 辛基酚聚氧乙烯醚磺酸盐界面行为的分子动力学模拟[J]. 化工学报, 2016, 67(4): 1416-1423. |
| [12] | 程亮, 徐丽, 侯翠红, 雒廷亮, 张保林, 刘国际. 低温条件下纳米腐殖酸-尿素配合物的制备及表征[J]. 化工学报, 2015, 66(7): 2725-2736. |
| [13] | 钱建华, 潘晓娜, 张强, 刘琳. 2,5-二芳基-1,3,4-噻二唑衍生物的合成及缓蚀性能[J]. 化工学报, 2015, 66(7): 2737-2748. |
| [14] | 高宁, 王一超, 刘育红. 丙炔基双酚A醚硼聚合物热解过程的ReaxFF分子动力学模拟[J]. 化工学报, 2015, 66(4): 1557-1564. |
| [15] | 王燕1,葛喜慧2,张敏卿1,朱怀工3,张子建2,王明1. 费托合成高温油相产品中正构烃的分离[J]. 化工进展, 2014, 33(11): 2894-2898. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备 11010102001995号
京公网安备 11010102001995号