化工学报 ›› 2021, Vol. 72 ›› Issue (5): 2426-2435.DOI: 10.11949/0438-1157.20201283
收稿日期:2020-09-08
修回日期:2020-11-25
出版日期:2021-05-05
发布日期:2021-05-05
通讯作者:
田平芳
作者简介:王欣(1996—),女,硕士研究生,基金资助:
WANG Xin1( ),ZHAO Peng1,LI Qingyang2,TIAN Pingfang1(
),ZHAO Peng1,LI Qingyang2,TIAN Pingfang1( )
)
Received:2020-09-08
Revised:2020-11-25
Online:2021-05-05
Published:2021-05-05
Contact:
TIAN Pingfang
摘要:
半导体合成生物学是研究半导体技术与合成生物学之间协同作用的一门交叉学科。其涉及的活细胞-半导体材料杂合体系具有独特的能量和信号转导机制,不仅维持活细胞的代谢能力,而且保留半导体材料的光电学物理特性,在化工、通讯、计算、能源及医疗等领域具有广阔的应用前景。综述了半导体合成生物学在生物催化、智能生物传感以及新型DNA数据存储领域的最新研究进展,讨论了目前研究面临的技术难题及解决方案,旨在为合成生物学和半导体技术这两个影响化工发展的领域提供有价值的参考。
中图分类号:
王欣, 赵鹏, 李清扬, 田平芳. 半导体合成生物学的研究进展[J]. 化工学报, 2021, 72(5): 2426-2435.
WANG Xin, ZHAO Peng, LI Qingyang, TIAN Pingfang. Research advances in semiconductor synthetic biology[J]. CIESC Journal, 2021, 72(5): 2426-2435.
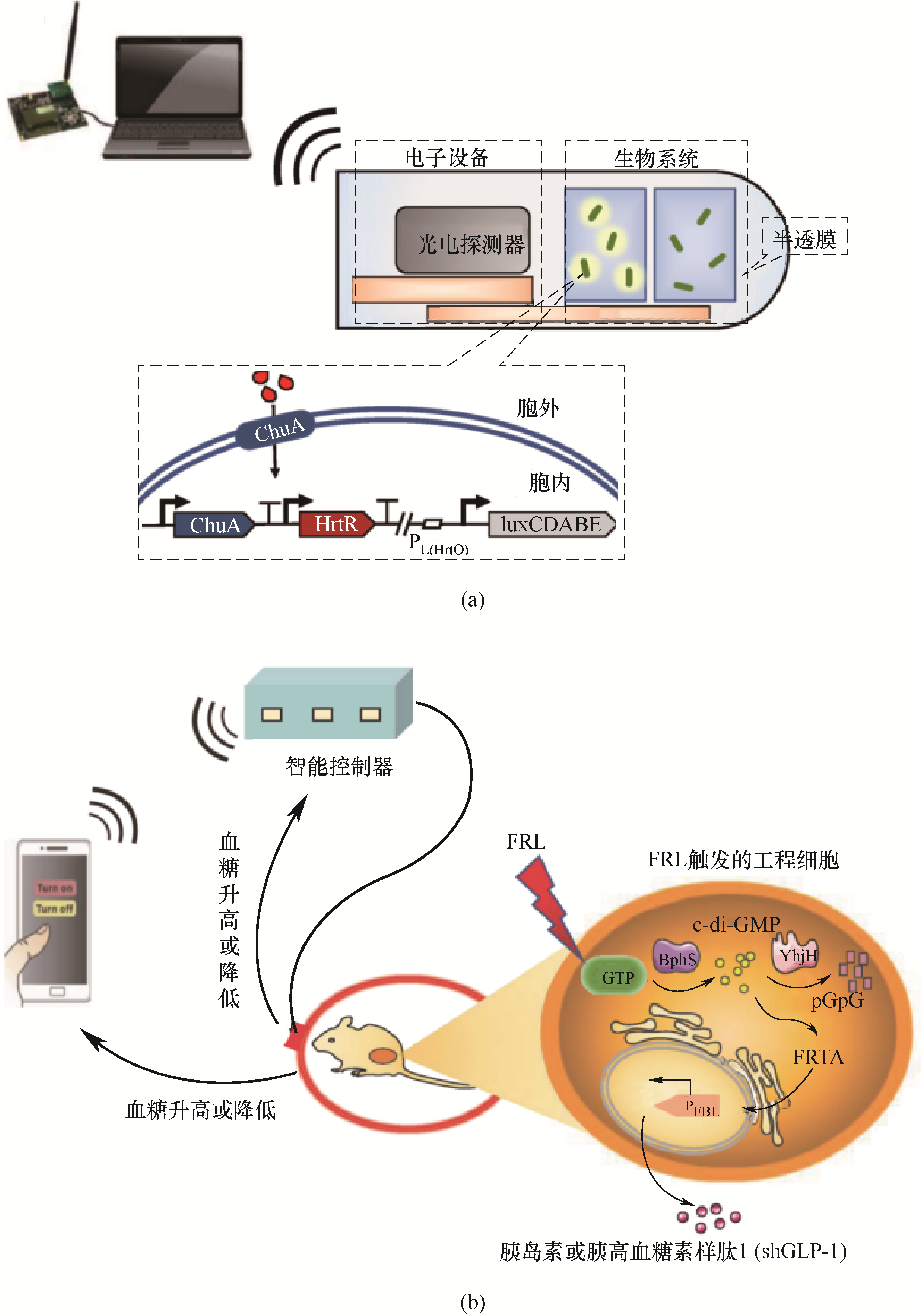
图2 可吸收的微电子设备(IMBED)示意图(a);智能手机调控工程细胞的表达,实现半自动血糖稳态 (b)ChuA—大肠杆菌外膜转运蛋白;HrtR—乳酸乳球菌血红素反应性转录抑制子;luxCDABE—荧光素酶的基因簇; FRL—远红光光源;BphS—基于细菌光活化的环二鸟苷酸合成酶;YhjH—c-di-GMP特异性磷酸二酯酶
Fig.2 Schematic diagram of absorbable microelectronic equipment (IMBED)(a); Smart phones regulate engineered cells to achieve semi-automatic blood glucose homeostasis (b)ChuA—outer membrane transporter of E. coli; HrtR—heme reactive transcriptional suppressor of Lactococcus lactis; luxCDABE—luciferase gene cluster; FRL—far-red light; BphS—bacterial light-activated cyclic diguanylate monophosphate (c-di-GMP) synthase; YhjH—c-di-GMP-specific phosphodiesterase
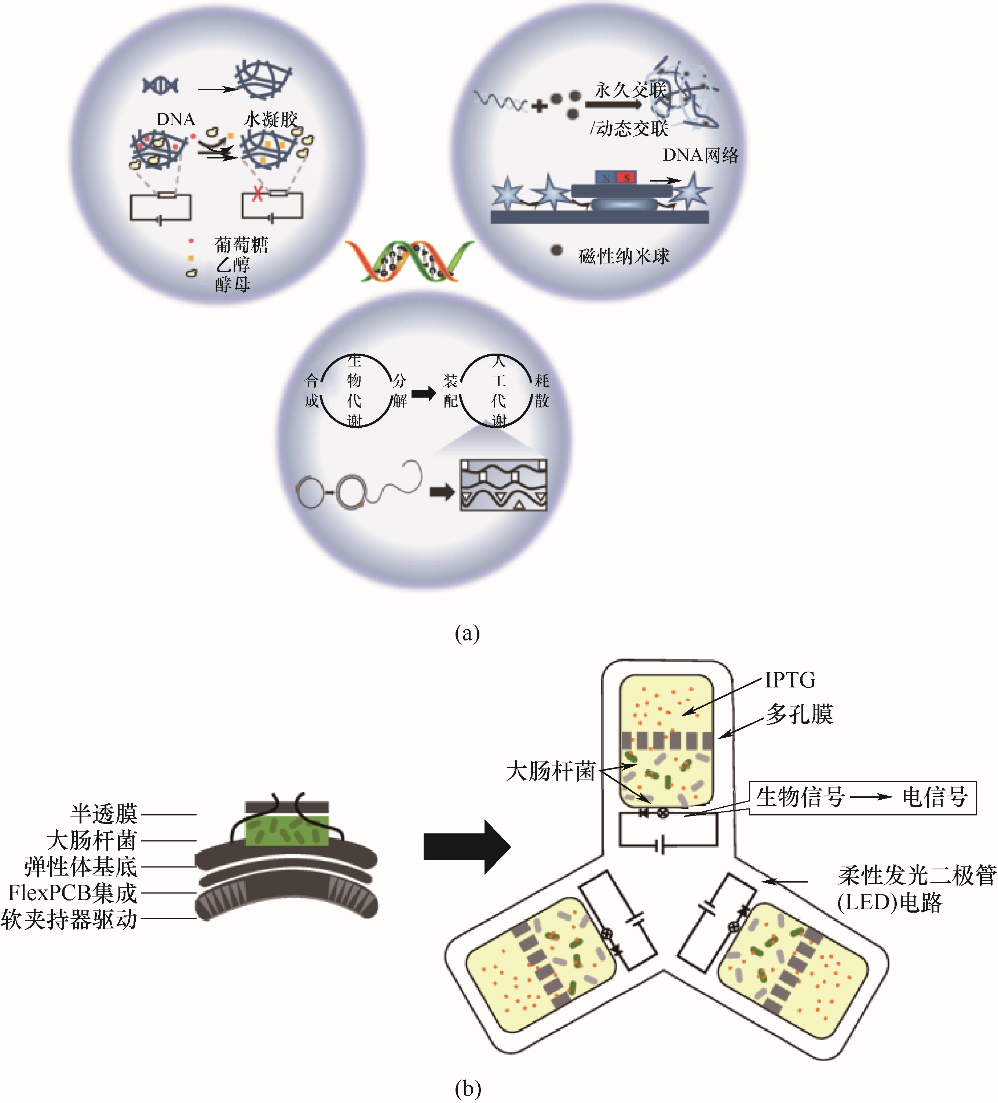
图3 DNA水凝胶的应用(a);工程细胞-柔性材料软机器人示意图 (b)
Fig.3 Applications of DNA hydrogel (a); Schematic diagram of engineered cell-flexible materials-based soft robot (b)
| 1 | Bartley B A, Kim K, Medley J K, et al. Synthetic biology: engineering living systems from biophysical principles [J]. Biophysical Journal, 2017, 112(6): 1050-1058. |
| 2 | Cheng A A, Lu T K. Synthetic biology: an emerging engineering discipline [J]. Annual Review of Biomedical Engineering, 2012, 14(1): 155-178. |
| 3 | Tian B, Xu S, Rogers J A, et al. Roadmap on semiconductor-cell biointerfaces [J]. Physical Biology, 2018, 15(3): 031002. |
| 4 | Fu W, Chaiboonchoe A, Khraiwesh B, et al. Intracellular spectral recompositioning of light enhances algal photosynthetic efficiency [J]. Sci. Adv., 2017, 3(9): e1603096. |
| 5 | Royanian S, Ziabari A A, Yousefi R. Efficiency enhancement of ultra-thin CIGS solar cells using bandgap grading and embedding Au plasmonic nanoparticles [J]. Plasmonics, 2020, 15: 1173-1182. |
| 6 | Shockley W, Queisser H J. Detailed balance limit of efficiency of p-n junction solar cells [J]. Journal of Applied Physics, 1961, 32(3): 510-519. |
| 7 | Zhang T, Tremblay P L. Hybrid photosynthesis-powering biocatalysts with solar energy captured by inorganic devices [J]. Biotechnology for Biofuels, 2017, 10(1): 249. |
| 8 | Claassens N J, Sousa D Z, dos Santos V A, et al. Harnessing the power of microbial autotrophy [J]. Nature Reviews Microbiology, 2016, 14(11): 692-706. |
| 9 | Dilek K D, Daniel G N. Artificial photosynthesis at efficiencies greatly exceeding that of natural photosynthesis [J]. Acc. Chem. Res., 2019, 52(11): 3143-3148. |
| 10 | Wei W, Sun P, Li Z, et al. A surface-display biohybrid approach to light-driven hydrogen production in air [J]. Sci. Adv., 2018, 4(2): eaap9253. |
| 11 | Guo J, Suastegui M, Sakimoto K K, et al. Light-driven fine chemical production in yeast biohybrids[J]. Science, 2018, 362(6416): 813-816. |
| 12 | Liu C, Gallagher J J, Sakimoto K K, et al. Nanowire-bacteria hybrids for unassisted solar carbon dioxide fixation to value-added chemicals [J]. Nano Lett., 2015, 15(5): 3634-3639. |
| 13 | Sakimoto K K, Wong A B, Yang P. Self-photosensitization of nonphotosynthetic bacteria for solar-to-chemical production [J]. Science, 2016, 351(6268): 74-77. |
| 14 | Xu M, Tremblay P L, Jiang L, et al. Stimulating bioplastic production with light energy by coupling Ralstonia eutropha with the photocatalyst graphitic carbon nitride [J]. Green Chemistry, 2019, 21(9): 2392-2400. |
| 15 | Tremblay P L, Xu M, Chen Y, et al. Nonmetallic abiotic-biological hybrid photocatalyst for visible water splitting and carbon dioxide reduction[J]. iScience, 2020, 23(1): 100784. |
| 16 | Kim W, Ng J K, Kunitake M E, et al. Interfacing silicon nanowires with mammalian cells [J]. Journal of the American Chemical Society, 2007, 129(23): 7228-7229. |
| 17 | 张宁, 徐开凯, 陈彦旭, 等.金属-氧化物-半导体硅发光器件在集成电路中的应用前景[J]. 物理学报, 2019, 68(16): 91-96. |
| Zhang N, Xu K K, Chen Y X, et al. Application prospect of metal-oxide-semiconductor silicon light emitting devices in integrated circuits [J]. Acta Physica Sinica, 2019, 68(16): 91-96. | |
| 18 | Brown K A, Harris D F, Wilker M B, et al. Light-driven dinitrogen reduction catalyzed by a CdS: nitrogenase MoFe protein biohybrid [J]. Science, 2016, 352(6284): 448-450. |
| 19 | Liu C, Colon B C, Ziesack M, et al. Water splitting-biosynthetic system with CO2 reduction efficiencies exceeding photosynthesis [J]. Science, 2016, 352(6290): 1210-1213. |
| 20 | Zhang H, Liu H, Tian Z, et al. Bacteria photosensitized by intracellular gold nanoclusters for solar fuel production[J]. Nat. Nanotechnol., 2018, 13(10): 900-905. |
| 21 | Sytnyk M, Jakesova M, Litvinukova M, et al. Cellular interfaces with hydrogen-bonded organic semiconductor hierarchical nanocrystals [J]. Nat. Commun., 2017, 8(1): 91. |
| 22 | Kracke F, Vassilev I, Kromer J O. Microbial electron transport and energy conservation-the foundation for optimizing bioelectrochemical systems [J]. Front. Microbiol., 2015, 6: 575. |
| 23 | Ye J, Yu J, Zhang Y, et al. Light-driven carbon dioxide reduction to methane by Methanosarcina barkeri-CdS biohybrid [J]. Applied Catalysis B: Environmental, 2019, 257: 117916. |
| 24 | Kornienko N, Sakimoto K K, Herlihy D M, et al. Spectroscopic elucidation of energy transfer in hybrid inorganic-biological organisms for solar-to-chemical production [J]. Proc. Natl. Acad. Sci. USA, 2016, 113(42): 11750-11755. |
| 25 | Cestellos-Blanco S, Zhang H, Kim J M, et al. Photosynthetic semiconductor biohybrids for solar-driven biocatalysis [J]. Nat. Catal., 2020, 3: 245-255. |
| 26 | Shi L, Dong H, Reguera G, et al. Extracellular electron transfer mechanisms between microorganisms and minerals [J]. Nat. Rev. Microbiol., 2016, 14(10): 651-662. |
| 27 | Jensen H M, Albers A E, Malley K R, et al. Engineering of a synthetic electron conduit in living cells [J]. Proc. Natl. Acad. Sci. USA, 2010, 107(45): 19213-19218. |
| 28 | Sakimoto K K, Kornienko N, Cestellos-Blanco S, et al. Physical biology of the materials-microorganism interface [J]. Journal of the American Chemical Society, 2018, 140(6): 1978-1985. |
| 29 | Gai P, Yu W, Zhao H, et al. Solar-powered organic semiconductor-bacteria biohybrids for CO2 reduction into acetic acid [J]. Angew. Chem. Int. Ed., 2020, 59(18): 7224-7229. |
| 30 | Suástegui M, Yu Ng C, Chowdhury A, et al. Multilevel engineering of the upstream module of aromatic amino acid biosynthesis in, Saccharomyces cerevisiae, for high production of polymer and drug precursors [J]. Metab. Eng., 2017, 42: 134-144. |
| 31 | Snyder P J, LaJeunesse D R, Reddy P, et al. Bioelectronics communication: encoding yeast regulatory responses using nanostructured gallium nitride thin films [J]. Nanoscale, 2018, 10: 11506–11516. |
| 32 | Kladko D V, Zakharzhevskii M A, Vinogradov V V. Magnetic field-mediated control of whole-cell biocatalysis [J]. J. Phys. Chem. Lett., 2020, 11: 8989-8996. |
| 33 | Updike S J, Hicks G P. The enzyme electrode [J]. Nature, 1967, 214(5092): 986-988. |
| 34 | Mimee M, Nadeau P, Hayward A, et al. An ingestible bacterial-electronic system to monitor gastrointestinal health [J]. Science, 2018, 360(6391): 915-918. |
| 35 | Shao J, Xue S, Yu G, et al. Smartphone-controlled optogenetically engineered cells enable semiautomatic glucose homeostasis in diabetic mice [J]. Sci. Transl. Med., 2017, 9(387): eaal2298. |
| 36 | Li F, Tang J, Geng J, et al. Polymeric DNA hydrogel: design, synthesis and applications [J]. Progress in Polymer Science, 2019, 98: 101163. |
| 37 | Han J, Cui Y, Han X, et al. Super-soft DNA/dopamine-grafted-dextran hydrogel as dynamic wire for electric circuits switched by a microbial metabolism process [J]. Adv. Sci. (Weinh), 2020, 7(13): 2000684. |
| 38 | Tang J, Yao C, Gu Z, et al. Super-soft and super-elastic DNA robot with magnetically driven navigational locomotion for cell delivery in confined space [J]. Angew. Chem. Int. Ed., 2020, 59(6): 2490-2495. |
| 39 | Hamada S, Yancey K G, Pardo Y, et al. Dynamic DNA material with emergent locomotion behavior powered by artificial metabolism [J]. Science Robotics, 2019, 4(29): eaaw3512. |
| 40 | Nawroth J C, Lee H, Feinberg A W, et al. A tissue-engineered jellyfish with biomimetic propulsion [J]. Nat. Biotechnol., 2012, 30(8): 792-797. |
| 41 | Cvetkovic C, Raman R, Chan V, et al. Three-dimensionally printed biological machines powered by skeletal muscle [J]. Proc. Natl. Acad. Sci. USA, 2014, 111(28): 10125-10130. |
| 42 | Feinberg A W, Feigel A, Shevkoplyas S S, et al. Muscular thin films for building actuators and powering devices[J]. Science, 2007, 317(5843): 1366-1370. |
| 43 | Justus K B, Hellebrekers T, Lewis D D, et al. A biosensing soft robot: autonomous parsing of chemical signals through integrated organic and inorganic interfaces [J]. Science Robotics, 2019, 4(31): eaax0765. |
| 44 | Strukov D B, Likharev K K. Defect-tolerant architectures for nanoelectronic crossbar memories [J]. Journal of Nanoscience and Nanotechnology, 2007, 7(1): 151-167. |
| 45 | Jiang Y, Tian B. Inorganic semiconductor biointerfaces[J]. Nat. Rev. Mater., 2018, 3(12): 473-490. |
| 46 | Allentoft M E, Collins M, Harker D, et al. The half-life of DNA in bone: measuring decay kinetics in 158 dated fossils [J]. Proc. Biol. Sci., 2012, 279(1748): 4724-4733. |
| 47 | Grass R N, Heckel R, Puddu M, et al. Robust chemical preservation of digital information on DNA in silica with error-correcting codes [J]. Angew. Chem. Int. Ed., 2015, 54(8): 2552-2555. |
| 48 | Bonnet J, Colotte M, Coudy D, et al. Chain and conformation stability of solid-state DNA: implications for room temperature storage [J]. Nucleic Acids Research, 2010, 38(5): 1531-1546. |
| 49 | Akram F, Haq I U, Ali H, et al. Trends to store digital data in DNA: an overview [J]. Mol. Biol. Rep., 2018, 45(5): 1479-1490. |
| 50 | Extance A. How DNA could store all the world's data [J]. Nature, 2016, 537(7618): 22-24. |
| 51 | Panda D, Molla K A, Baig M J, et al. DNA as a digital information storage device: hope or hype? [J]. 3 Biotech, 2018, 8(5): 239. |
| 52 | Church G M, Gao Y, Kosuri S. Next-generation digital information storage in DNA [J]. Science, 2012, 337(6102): 1628. |
| 53 | Greenberg A, Hamilton J, Maltz D A, et al. The cost of a cloud: research problems in data center networks [J]. Acm Sigcomm Computer Communication Review, 2008, 39(1): 68-73. |
| 54 | de Silva P Y, Ganegoda G U. New trends of digital data storage in DNA [J]. Biomed. Res. Int., 2016, 2016: 8072463. |
| 55 | Ceze L, Nivala J, Strauss K. Molecular digital data storage using DNA [J]. Nature Reviews Genetics, 2019, 20(8): 456-466. |
| 56 | Ping Z, Ma D, Huang X, et al. Carbon-based archiving: current progress and future prospects of DNA-based data storage [J]. Gigascience, 2019, 8(6): giz075. |
| 57 | Schwartz J J, Lee C, Shendure J. Accurate gene synthesis with tag-directed retrieval of sequence-verified DNA molecules [J]. Nature Methods, 2012, 9(9): 913-915. |
| 58 | Erlich Y, Zielinski D. DNA fountain enables a robust and efficient storage architecture [J]. Science, 2017, 355(6328): 950-954. |
| 59 | Huffman D A. A method for the construction of minimum-redundancy codes [J]. Resonance, 2006, 11(2): 91-99. |
| 60 | Blawat M, Gaedke K, Huetter I, et al. Forward error correction for DNA data storage [J]. Procedia Computer Science, 2016, 80: 1011-1022. |
| 61 | Goldman N, Bertone P, Chen S, et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA [J]. Nature, 2013, 494(7435): 77-80. |
| 62 | Bornholt J, Lopez R, Carmean D, et al. A DNA-based archival storage system [J]. ACM SIGPLAN Notices, 2016, 51(4): 637-649. |
| 63 | Baum E B. Building an associative memory vastly larger than the brain [J]. Science, 1995, 268(5210): 583-585. |
| 64 | Yazdi S M, Yuan Y, Ma J, et al. A rewritable, random-access DNA-based storage system [J]. Sci. Rep., 2015, 5: 14138. |
| 65 | Organick L, Ang S D, Chen Y J, et al. Random access in large-scale DNA data storage [J]. Nature Biotechnology, 2018, 36: 242-248. |
| 66 | Goodwin S, Mcpherson J D, Mccombie W R. Coming of age: ten years of next-generation sequencing technologies [J]. Nat. Rev. Genet., 2016, 17(6): 333-351. |
| 67 | Shendure J, Balasubramanian S, Church G M, et al. DNA sequencing at 40: past, present and future [J]. Nature, 2017, 550(7676): 345-353. |
| 68 | Deamer D, Akeson M, Branton D. Three decades of nanopore sequencing [J]. Nature Biotechnology, 2016, 34(5): 518-524. |
| 69 | Shipman S L, Nivala J, Macklis J D, et al. CRISPR-Cas encoding of a digital movie into the genomes of a population of living bacteria [J]. Nature, 2017, 547(7663): 345-349. |
| 70 | Jain S, Farnoud F, Schwartz M, et al. Duplication-correcting codes for data storage in the DNA of living organisms [J]. IEEE Transactions on Information Theory, 2017, 63(8): 4996-5010. |
| 71 | Nguyen H H, Park J, Park S J, et al. Long-term stability and integrity of plasmid-based DNA data storage [J]. Polymers (Basel), 2018, 10(1): 28. |
| 72 | Tian B, Lieber C M. Nanowired bioelectric interfaces [J]. Chem. Rev., 2019, 119(15): 9136-9152. |
| [1] | 杨松杰, 田禾. 有机光致变色材料最新研究进展 [J]. 化工学报, 2003, 54(4): 497-507. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备 11010102001995号
京公网安备 11010102001995号