CIESC Journal ›› 2024, Vol. 75 ›› Issue (4): 1655-1667.DOI: 10.11949/0438-1157.20230992
• Process system engineering • Previous Articles Next Articles
Huaqiang WEN( ), Quanhu SUN(
), Quanhu SUN( ), Weifeng SHEN(
), Weifeng SHEN( )
)
Received:2023-09-21
Revised:2023-12-19
Online:2024-06-06
Published:2024-04-25
Contact:
Weifeng SHEN
通讯作者:
申威峰
作者简介:文华强(1997—),男,博士研究生,huaqiangwen@cqu.edu.cn基金资助:CLC Number:
Huaqiang WEN, Quanhu SUN, Weifeng SHEN. Targeted intelligent molecular generation framework based on fragments chemical space[J]. CIESC Journal, 2024, 75(4): 1655-1667.
文华强, 孙全虎, 申威峰. 基于分子碎片化学空间的智能分子定向生成框架[J]. 化工学报, 2024, 75(4): 1655-1667.
Add to citation manager EndNote|Ris|BibTeX
| 筛选项目 | 原始范围 | 最佳值 | 容忍范围 |
|---|---|---|---|
| QED | 0~1 | 1 | 0.8~1 |
| SAscore | 1~10 | 1 | 1~3 |
| SlogP | — | 1.5 | 0~3 |
| IDP | 0~1.5 | 0 | 0~0.8 |
Table 1 Criteria for screening excellent molecules
| 筛选项目 | 原始范围 | 最佳值 | 容忍范围 |
|---|---|---|---|
| QED | 0~1 | 1 | 0.8~1 |
| SAscore | 1~10 | 1 | 1~3 |
| SlogP | — | 1.5 | 0~3 |
| IDP | 0~1.5 | 0 | 0~0.8 |
| 指纹类别 | 指纹长度 | 学习最大迭代次数 | 学习率 | Neighborhood function | Sigma | 特征初始化算法 | 随机种子 |
|---|---|---|---|---|---|---|---|
| RDKit Topological | 1024 | 1000 | 0.5 | ‘gaussian’ | 3 | 主成分分析降维 | 2023 |
Table 2 Parameter of SOM clustering network based on Topological molecular fingerprints
| 指纹类别 | 指纹长度 | 学习最大迭代次数 | 学习率 | Neighborhood function | Sigma | 特征初始化算法 | 随机种子 |
|---|---|---|---|---|---|---|---|
| RDKit Topological | 1024 | 1000 | 0.5 | ‘gaussian’ | 3 | 主成分分析降维 | 2023 |
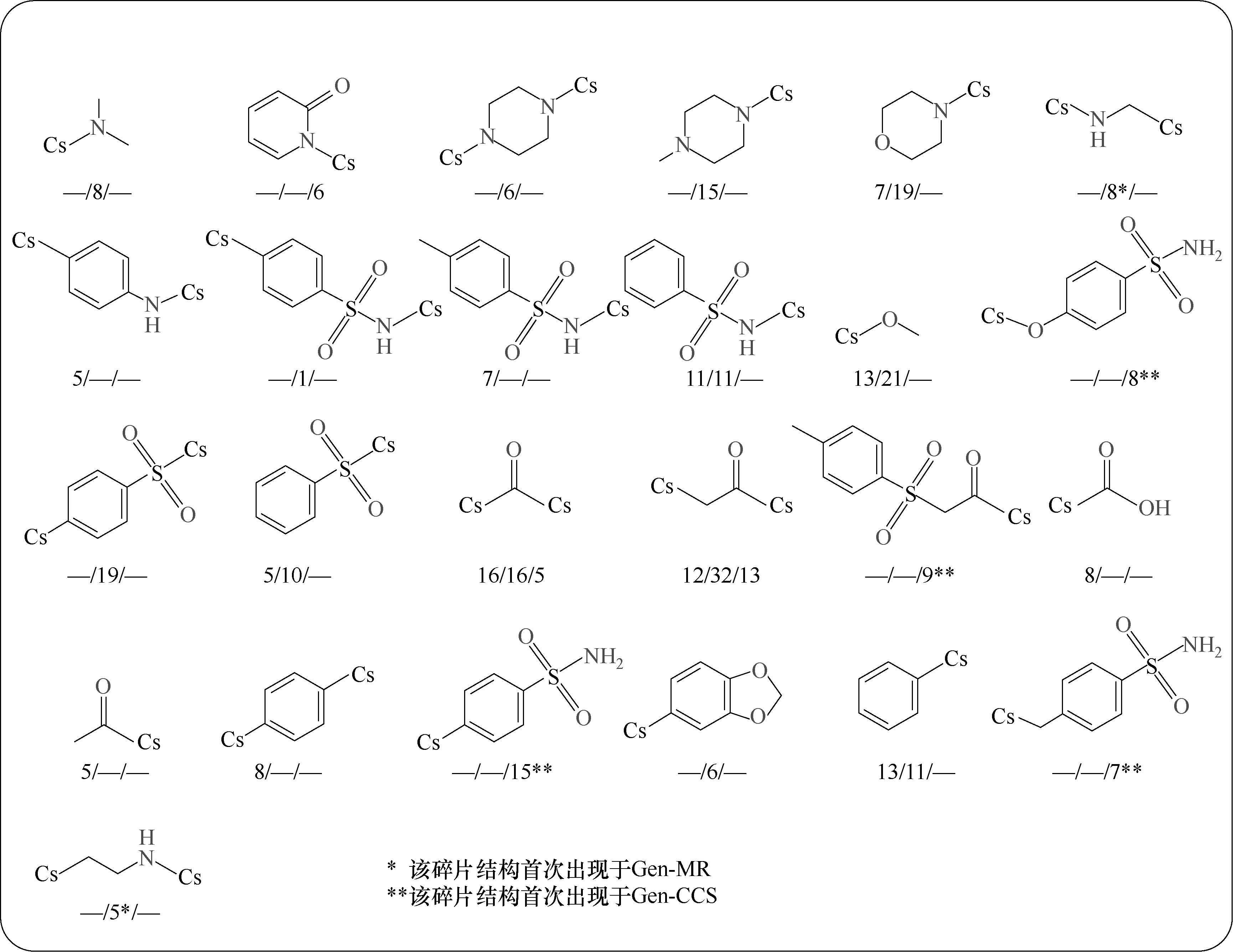
Fig.10 Fragment structure and frequency statistics of excellent molecular clusters (IDP <0.6) in Gen0, Gen-MR, and Gen-CSS (the displayed fragment frequency is not less than 5, and it is distributed as “frequency in Gen0/frequency in Gen-MR/frequency in Gen-CSS”)
| 1 | Sanchez-Lengeling B, Aspuru-Guzik A. Inverse molecular design using machine learning: generative models for matter engineering[J]. Science, 2018, 361(6400): 360-365. |
| 2 | Chen H M, Engkvist O, Wang Y H, et al. The rise of deep learning in drug discovery[J]. Drug Discovery Today, 2018, 23(6): 1241-1250. |
| 3 | Butler K T, Davies D W, Cartwright H, et al. Machine learning for molecular and materials science[J]. Nature, 2018, 559(7715): 547-555. |
| 4 | Lu S H, Zhou Q H, Ouyang Y X, et al. Accelerated discovery of stable lead-free hybrid organic-inorganic perovskites via machine learning[J]. Nature Communications, 2018, 9: 3405. |
| 5 | Pretel E J, López P A, Bottini S B, et al. Computer-aided molecular design of solvents for separation processes[J]. AIChE Journal, 1994, 40(8): 1349-1360. |
| 6 | Scheffczyk J, Fleitmann L, Schwarz A, et al. COSMO-CAMD: a framework for optimization-based computer-aided molecular design using COSMO-RS[J]. Chemical Engineering Science, 2017, 159: 84-92. |
| 7 | 赵红庆, 刘奇磊, 张磊, 等. 考虑选择性和反应速率的多目标制药反应溶剂设计[J]. 化工学报, 2021, 72(3): 1465-1472. |
| Zhao H Q, Liu Q L, Zhang L, et al. Multi-objective solvent design considering selectivity and reaction rate for pharmaceutical reactions[J]. CIESC Journal, 2021, 72(3): 1465-1472. | |
| 8 | Gani R, Nielsen B, Fredenslund A. A group contribution approach to computer-aided molecular design[J]. AIChE Journal, 1991, 37(9): 1318-1332. |
| 9 | 张学岗, 张军保, 宋静, 等. 基于MGASA的计算机辅助分子设计[J]. 化工进展, 2008, 27(12): 2019-2024. |
| Zhang X G, Zhang J B, Song J, et al. Computer aided molecular design based on MGASA[J]. Chemical Industry and Engineering Progress, 2008, 27(12): 2019-2024. | |
| 10 | von Lilienfeld O A, Müller K R, Tkatchenko A. Exploring chemical compound space with quantum-based machine learning[J]. Nature Reviews Chemistry, 2020, 4(7): 347-358. |
| 11 | Hochreiter S, Schmidhuber J. Long short-term memory[J]. Neural Computation, 1997, 9(8): 1735-1780. |
| 12 | Kingma D P, Welling M. Auto-encoding variational bayes[EB/OL]. 2014, . |
| 13 | Goodfellow I, Pouget-Abadie J, Mirza M, et al. Generative adversarial networks[J]. Communications of the ACM, 2020, 63(11): 139-144. |
| 14 | Kusner M J, Paige B, Hernández-Lobato J M. Grammar variational autoencoder[C]//Precup D, Teh Y W. Proceedings of the 34th International Conference on Machine Learning. Sydney, Australia, 2017: 1945-1954. |
| 15 | Grisoni F, Moret M, Lingwood R, et al. Bidirectional molecule generation with recurrent neural networks[J]. Journal of Chemical Information and Modeling, 2020, 60(3): 1175-1183. |
| 16 | Gómez-Bombarelli R, Wei J N, Duvenaud D, et al. Automatic chemical design using a data-driven continuous representation of molecules[J]. ACS Central Science, 2018, 4(2): 268-276. |
| 17 | Sánchez-Lengeling B, Outeiral C, Guimaraes G, et al. Optimizing distributions over molecular space. An objective-reinforced generative adversarial network for inverse-design chemistry(ORGANIC)[EB/OL]. Cambridge: Cambridge Open Engage, 2017, . |
| 18 | Jin W G, Barzilay R, Jaakkola T. Junction tree variational autoencoder for molecular graph generation[EB/OL]. 2018, . |
| 19 | Bagal V, Aggarwal R, Vinod P K, et al. MolGPT: molecular generation using a transformer-decoder model[J]. Journal of Chemical Information and Modeling, 2022, 62(9): 2064-2076. |
| 20 | Barredo Arrieta A, Díaz-Rodríguez N, Del Ser J, et al. Explainable artificial intelligence (XAI): concepts, taxonomies, opportunities and challenges toward responsible AI[J]. Information Fusion, 2020, 58: 82-115. |
| 21 | Degen J, Wegscheid-Gerlach C, Zaliani A, et al. On the art of compiling and using ‘drug-like’ chemical fragment spaces[J]. ChemMedChem, 2008, 3(10): 1503-1507. |
| 22 | Kim S, Thiessen P A, Bolton E E, et al. PubChem substance and compound databases[J]. Nucleic Acids Research, 2016, 44(D1): D1202-D1213. |
| 23 | Kohonen T. Self-organized formation of topologically correct feature maps[J]. Biological Cybernetics, 1982, 43(1): 59-69. |
| 24 | Gaulton A, Bellis L J, Bento A P, et al. ChEMBL: a large-scale bioactivity database for drug discovery[J]. Nucleic Acids Research, 2012, 40(D1): D1100-D1107. |
| 25 | SMILES Weininger D., a chemical language and information system( 1): Introduction to methodology and encoding rules[J]. Journal of Chemical Information and Computer Sciences, 1988, 28(1): 31-36. |
| 26 | Nilakantan R, Bauman N, Dixon J S, et al. Topological torsion: a new molecular descriptor for SAR applications. Comparison with other descriptors[J]. Journal of Chemical Information and Computer Sciences, 1987, 27(2): 82-85. |
| 27 | Durant J L, Leland B A, Henry D R, et al. Reoptimization of MDL keys for use in drug discovery[J]. Journal of Chemical Information and Computer Sciences, 2002, 42(6): 1273-1280. |
| 28 | Landrum G. RDKit: open-source cheminformatics software (version 2021.09.1)[CP/OL]. [2023-06-15]. . |
| 29 | Ertl P, Schuffenhauer A. Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions[J]. Journal of Cheminformatics, 2009, 1(1): 8. |
| 30 | Bickerton G R, Paolini G V, Besnard J, et al. Quantifying the chemical beauty of drugs[J]. Nature Chemistry, 2012, 4(2): 90-98. |
| 31 | Wildman S A, Crippen G M. Prediction of physicochemical parameters by atomic contributions[J]. Journal of Chemical Information and Computer Sciences, 1999, 39(5): 868-873. |
| 32 | Hughes J D, Blagg J, Price D A, et al. Physiochemical drug properties associated with in vivo toxicological outcomes[J]. Bioorganic & Medicinal Chemistry Letters, 2008, 18(17): 4872-4875. |
| [1] | Kaijie WEN, Li GUO, Zhaojie XIA, Jianhua CHEN. A rapid simulation method of gas-solid flow by coupling CFD and deep learning [J]. CIESC Journal, 2023, 74(9): 3775-3785. |
| [2] | Shuping QI, Wenlong WANG, Lei ZHANG, Jian DU. A deep learning-based model for predicting the stability constants of metal ions with organic ligands [J]. CIESC Journal, 2022, 73(12): 5461-5468. |
| [3] | XIE Haoyuan, HUANG Qunxing, LIN Xiaoqing, LI Xiaodong, YAN Jianhua. Study on the calorific value prediction of municipal solid wastes by image deep learning [J]. CIESC Journal, 2021, 72(5): 2773-2782. |
| [4] | CHEN Zhongsheng, ZHU Meiyu, HE Yanlin, XU Yuan, ZHU Qunxiong. Quantile regression CGAN based virtual samples generation and its applications to process modeling [J]. CIESC Journal, 2021, 72(3): 1529-1538. |
| [5] | YU Chengyuan, WU Jinkui, ZHOU Li, JI Xu, DAI Yiyang, DANG Yagu. Prediction of energy conversion efficiency of organic solar cells based on deep learning [J]. CIESC Journal, 2021, 72(3): 1487-1495. |
| [6] | Xiaohui WANG, Yanjiang WANG, Xiaogang DENG, Zheng ZHANG. Industrial process fault detection using weighted deep support vector data description [J]. CIESC Journal, 2021, 72(11): 5707-5716. |
| [7] | Linzi YIN, Yuyin GUAN, Zhaohui JIANG, Xuemei XU. Optimal method of selecting silicon content data in blast furnace hot metal based on k-means++ [J]. CIESC Journal, 2020, 71(8): 3661-3670. |
| [8] | Luyao TIAN, Zihao WANG, Yang SU, Huaqiang WEN, Weifeng SHEN. Research advances in deep learning based quantitative structure-property relationship modeling of solvents [J]. CIESC Journal, 2020, 71(10): 4462-4472. |
| [9] | Hengchang GU, Peng MU, Jianwei LI. Modeling and application of ethylene cracking furnace based on cross-iterative BLSTM network [J]. CIESC Journal, 2019, 70(2): 548-555. |
| [10] | DONG Shun, LI Yiguo, SUN Shuanzhu, LIU Xichui, SHEN Jiong. Fault detection method based on state space-PCANet [J]. CIESC Journal, 2018, 69(8): 3528-3536. |
| [11] | WANG Kangcheng, SHANG Chao, KE Wensi, JIANG Yongheng, HUANG Dexian. Automatic structure and parameters tuning method for deep neural network soft sensor in chemical industries [J]. CIESC Journal, 2018, 69(3): 900-906. |
| [12] | WANG Gongming, LI Wenjing, QIAO Junfei. Prediction of effluent total phosphorus using PLSR-based adaptive deep belief network [J]. CIESC Journal, 2017, 68(5): 1987-1997. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||