化工学报 ›› 2024, Vol. 75 ›› Issue (11): 4141-4151.DOI: 10.11949/0438-1157.20240602
收稿日期:2024-06-03
修回日期:2024-10-01
出版日期:2024-11-25
发布日期:2024-12-26
通讯作者:
吉远辉
作者简介:丁叶薇(2000—),女,硕士研究生,dingyewei@seu.edu.cn
基金资助:
Yewei DING( ), Wenbo KANG, Yutong SONG, Qinxi FAN, Yuanhui JI(
), Wenbo KANG, Yutong SONG, Qinxi FAN, Yuanhui JI( )
)
Received:2024-06-03
Revised:2024-10-01
Online:2024-11-25
Published:2024-12-26
Contact:
Yuanhui JI
摘要:
以吲哚美辛作为模型药物,选用不同种类的抗肿瘤药物探究其与吲哚美辛形成自组装纳米粒的能力,采用Hansen溶解度参数模型、COSMO-RS理论模型、结合能计算等不同手段获得描述符预测纳米自组装行为与基于分子指纹作为描述符的机器学习算法预测进行对比验证,并通过分子动力学模拟可视化分子自组装过程以及量化计算进行分子间相互作用能分析发现氢键为自组装过程关键驱动力。研究表明,机器学习算法可快速预测药物分子自聚集和与吲哚美辛分子共聚集的概率初步筛选出药物分子组合,进一步结合热力学理论模型研究比较药物分子自身与不同药物分子间结合能大小、Hansen溶解度参数差值以及表面电荷密度分布等描述符可更准确地筛选合适的药物组合,为设计制备递送效率高、可联合治疗的无载体纳米药物递送系统提供重要的理论指导。
中图分类号:
丁叶薇, 康文博, 宋昱潼, 樊钦习, 吉远辉. 吲哚美辛纳米药物筛选及自组装机制的理论研究[J]. 化工学报, 2024, 75(11): 4141-4151.
Yewei DING, Wenbo KANG, Yutong SONG, Qinxi FAN, Yuanhui JI. Mechanism and screening of indomethacin self-assembled nanomedical drugs[J]. CIESC Journal, 2024, 75(11): 4141-4151.
| 类别 | 药物名称及缩写 | SMILES结构编码 | ||
|---|---|---|---|---|
| 非甾体抗炎药 | 吲哚美辛 | Indomethacin | IND | CC1=C(C2=C(N1C(=O)C3=CC=C(C=C3)Cl)C=CC(=C2)OC)CC(=O)O |
| 抗代谢类药物 | 氟达拉滨 | Fludarabine | FD | C1=NC2=C(N=C(N=C2N1C3C(C(C(O3)CO)O)O)F)N |
| 卡培他滨 | Capecitabine | CA | CCCCCOC(=O)NC1=NC(=O)N(C=C1F)[C@@H]1O[C@H](C)[C@@H](O)[C@H]1O | |
| 抗生素类 | 柔红霉素 | Daunorubicin | DA | COC1=CC=CC2=C1C(=O)C1=C(O)C3=C(C[C@](O)(C[C@@H]3O[C@H]3C[C@H](N)[C@H](O)[C@H](C)O3)C(C)=O)C(O)=C1C2=O |
| 阿霉素 | Adriamycin | AD | COC1=CC=CC2=C1C(=O)C1=C(O)C3=C(C[C@](O)(C[C@@H]3O[C@H]3C[C@H](N)[C@H](O)[C@H](C)O3)C(=O)CO)C(O)=C1C2=O | |
| 拓扑异构酶类 | 伊立替康 | Irinotecan | IR | CCC1=C2CN3C(=CC4=C(COC(=O)[C@]4(O)CC)C3=O)C2=NC2=CC=C(OC(=O)N3CCC(CC3)N3CCCCC3)C=C12 |
| 拓扑替康 | Topotecan | TP | CC[C@@]1(C2=C(COC1=O)C(=O)N3CC4=CC5=C(C=CC(=C5CN(C)C)O)N=C4C3=C2)O | |
| 激素类 | 他莫昔芬 | Tamoxifen | TF | CC/C(=C(\C1=CC=CC=C1)/C2=CC=C(C=C2)OCCN(C)C)/C3=CC=CC=C3 |
| 阿那曲唑 | Anastrozole | AZ | CC(C)(C#N)C1=CC(=CC(=C1)CN2C=NC=N2)C(C)(C)C#N | |
| 依西美坦 | Exemestane | ET | C[C@]12CC[C@H]3[C@H]([C@@H]1CCC2=O)CC(=C)C4=CC(=O)C=C[C@]34C | |
| 血管内皮生长 因子受体类 | 帕唑帕尼 | Pazopanib | PB | CC1=C(C=C(C=C1)NC2=NC=CC(=N2)N(C)C3=CC4=NN(C(=C4C=C3)C)C)S(=O)(=O)N |
| 阿西替尼 | Axitinib | AX | CNC(=O)C1=CC=CC=C1SC2=CC3=C(C=C2)C(=NN3)/C=C/C4=CC=CC=N4 | |
| 蛋白酶体 抑制剂类 | 硼替佐米 | Bortezomib | BZ | B([C@H](CC(C)C)NC(=O)[C@H](CC1=CC=CC=C1)NC(=O)C2=NC=CN=C2)(O)O |
| 转谷氨酰胺酶2 (TGase2)抑制剂 | GK921 | GK | C1CCN(C1)CCOC2=NC3=C(N=CC=C3)N=C2C#CC4=CC=CC=C4 | |
| 组蛋白去乙酰化酶类 | 贝利司他 | Belinostat | BE | ONC(=O)\C=C\C1=CC=CC(=C1)S(=O)(=O)NC1=CC=CC=C1 |
表1 本文中选择研究的药物
Table 1 The drugs selected in this paper
| 类别 | 药物名称及缩写 | SMILES结构编码 | ||
|---|---|---|---|---|
| 非甾体抗炎药 | 吲哚美辛 | Indomethacin | IND | CC1=C(C2=C(N1C(=O)C3=CC=C(C=C3)Cl)C=CC(=C2)OC)CC(=O)O |
| 抗代谢类药物 | 氟达拉滨 | Fludarabine | FD | C1=NC2=C(N=C(N=C2N1C3C(C(C(O3)CO)O)O)F)N |
| 卡培他滨 | Capecitabine | CA | CCCCCOC(=O)NC1=NC(=O)N(C=C1F)[C@@H]1O[C@H](C)[C@@H](O)[C@H]1O | |
| 抗生素类 | 柔红霉素 | Daunorubicin | DA | COC1=CC=CC2=C1C(=O)C1=C(O)C3=C(C[C@](O)(C[C@@H]3O[C@H]3C[C@H](N)[C@H](O)[C@H](C)O3)C(C)=O)C(O)=C1C2=O |
| 阿霉素 | Adriamycin | AD | COC1=CC=CC2=C1C(=O)C1=C(O)C3=C(C[C@](O)(C[C@@H]3O[C@H]3C[C@H](N)[C@H](O)[C@H](C)O3)C(=O)CO)C(O)=C1C2=O | |
| 拓扑异构酶类 | 伊立替康 | Irinotecan | IR | CCC1=C2CN3C(=CC4=C(COC(=O)[C@]4(O)CC)C3=O)C2=NC2=CC=C(OC(=O)N3CCC(CC3)N3CCCCC3)C=C12 |
| 拓扑替康 | Topotecan | TP | CC[C@@]1(C2=C(COC1=O)C(=O)N3CC4=CC5=C(C=CC(=C5CN(C)C)O)N=C4C3=C2)O | |
| 激素类 | 他莫昔芬 | Tamoxifen | TF | CC/C(=C(\C1=CC=CC=C1)/C2=CC=C(C=C2)OCCN(C)C)/C3=CC=CC=C3 |
| 阿那曲唑 | Anastrozole | AZ | CC(C)(C#N)C1=CC(=CC(=C1)CN2C=NC=N2)C(C)(C)C#N | |
| 依西美坦 | Exemestane | ET | C[C@]12CC[C@H]3[C@H]([C@@H]1CCC2=O)CC(=C)C4=CC(=O)C=C[C@]34C | |
| 血管内皮生长 因子受体类 | 帕唑帕尼 | Pazopanib | PB | CC1=C(C=C(C=C1)NC2=NC=CC(=N2)N(C)C3=CC4=NN(C(=C4C=C3)C)C)S(=O)(=O)N |
| 阿西替尼 | Axitinib | AX | CNC(=O)C1=CC=CC=C1SC2=CC3=C(C=C2)C(=NN3)/C=C/C4=CC=CC=N4 | |
| 蛋白酶体 抑制剂类 | 硼替佐米 | Bortezomib | BZ | B([C@H](CC(C)C)NC(=O)[C@H](CC1=CC=CC=C1)NC(=O)C2=NC=CN=C2)(O)O |
| 转谷氨酰胺酶2 (TGase2)抑制剂 | GK921 | GK | C1CCN(C1)CCOC2=NC3=C(N=CC=C3)N=C2C#CC4=CC=CC=C4 | |
| 组蛋白去乙酰化酶类 | 贝利司他 | Belinostat | BE | ONC(=O)\C=C\C1=CC=CC(=C1)S(=O)(=O)NC1=CC=CC=C1 |
| δd | δp | δh | δ | Δδ | δV | Δδd | Δδp | Δδh | Ra | |
|---|---|---|---|---|---|---|---|---|---|---|
| IND | 20.7 | 6.5 | 6.8 | 22.7 | 0 | 21.70 | 0 | 0 | 0 | 0 |
| FD | 18.7 | 14.4 | 16.4 | 28.8 | 6.1 | 23.60 | 2.0 | 7.9 | 9.6 | 14.78 |
| CA | 17.4 | 10.5 | 8.5 | 22.0 | 0.7 | 20.32 | 3.3 | 4.0 | 1.7 | 13.90 |
| DA | 20.7 | 12.6 | 7.8 | 25.5 | 2.8 | 24.23 | 0 | 6.1 | 1.0 | 6.18 |
| AD | 20.8 | 13.6 | 7.4 | 25.9 | 3.2 | 24.85 | 0.1 | 7.1 | 0.6 | 7.14 |
| IR | 20.7 | 4.1 | 7.7 | 22.4 | 0.3 | 21.10 | 0 | 2.4 | 0.9 | 2.56 |
| TP | 20.0 | 3.1 | 10.8 | 23.0 | 0.3 | 20.24 | 0.7 | 3.4 | 4.0 | 5.95 |
| TF | 18.4 | 2.9 | 3.6 | 19.0 | 3.7 | 18.63 | 2.3 | 3.6 | 3.2 | 10.38 |
| AZ | 18.0 | 11.1 | 3.8 | 21.5 | 1.2 | 21.15 | 2.7 | 4.6 | 3.0 | 12.12 |
| ET | 18.9 | 5.4 | 2.8 | 19.9 | 2.8 | 19.66 | 1.8 | 1.1 | 4.0 | 8.31 |
| PB | 21.5 | 10.4 | 7.9 | 25.1 | 2.4 | 23.88 | 0.8 | 3.9 | 1.1 | 5.16 |
| AX | 22.4 | 11.1 | 6.2 | 25.8 | 3.1 | 25.00 | 1.7 | 4.6 | 0.6 | 8.23 |
| BZ | 19.4 | 11.0 | 18.5 | 29.0 | 6.3 | 22.30 | 1.3 | 4.5 | 11.7 | 13.57 |
| GK | 19.1 | 6.6 | 4.8 | 20.8 | 1.9 | 20.21 | 1.6 | 0.1 | 2.0 | 6.71 |
| BE | 20.9 | 14.9 | 15.9 | 30.2 | 7.5 | 25.67 | 0.2 | 8.4 | 9.1 | 12.41 |
表2 Hansen溶解度参数值
Table 2 Hansen solubility parameters
| δd | δp | δh | δ | Δδ | δV | Δδd | Δδp | Δδh | Ra | |
|---|---|---|---|---|---|---|---|---|---|---|
| IND | 20.7 | 6.5 | 6.8 | 22.7 | 0 | 21.70 | 0 | 0 | 0 | 0 |
| FD | 18.7 | 14.4 | 16.4 | 28.8 | 6.1 | 23.60 | 2.0 | 7.9 | 9.6 | 14.78 |
| CA | 17.4 | 10.5 | 8.5 | 22.0 | 0.7 | 20.32 | 3.3 | 4.0 | 1.7 | 13.90 |
| DA | 20.7 | 12.6 | 7.8 | 25.5 | 2.8 | 24.23 | 0 | 6.1 | 1.0 | 6.18 |
| AD | 20.8 | 13.6 | 7.4 | 25.9 | 3.2 | 24.85 | 0.1 | 7.1 | 0.6 | 7.14 |
| IR | 20.7 | 4.1 | 7.7 | 22.4 | 0.3 | 21.10 | 0 | 2.4 | 0.9 | 2.56 |
| TP | 20.0 | 3.1 | 10.8 | 23.0 | 0.3 | 20.24 | 0.7 | 3.4 | 4.0 | 5.95 |
| TF | 18.4 | 2.9 | 3.6 | 19.0 | 3.7 | 18.63 | 2.3 | 3.6 | 3.2 | 10.38 |
| AZ | 18.0 | 11.1 | 3.8 | 21.5 | 1.2 | 21.15 | 2.7 | 4.6 | 3.0 | 12.12 |
| ET | 18.9 | 5.4 | 2.8 | 19.9 | 2.8 | 19.66 | 1.8 | 1.1 | 4.0 | 8.31 |
| PB | 21.5 | 10.4 | 7.9 | 25.1 | 2.4 | 23.88 | 0.8 | 3.9 | 1.1 | 5.16 |
| AX | 22.4 | 11.1 | 6.2 | 25.8 | 3.1 | 25.00 | 1.7 | 4.6 | 0.6 | 8.23 |
| BZ | 19.4 | 11.0 | 18.5 | 29.0 | 6.3 | 22.30 | 1.3 | 4.5 | 11.7 | 13.57 |
| GK | 19.1 | 6.6 | 4.8 | 20.8 | 1.9 | 20.21 | 1.6 | 0.1 | 2.0 | 6.71 |
| BE | 20.9 | 14.9 | 15.9 | 30.2 | 7.5 | 25.67 | 0.2 | 8.4 | 9.1 | 12.41 |
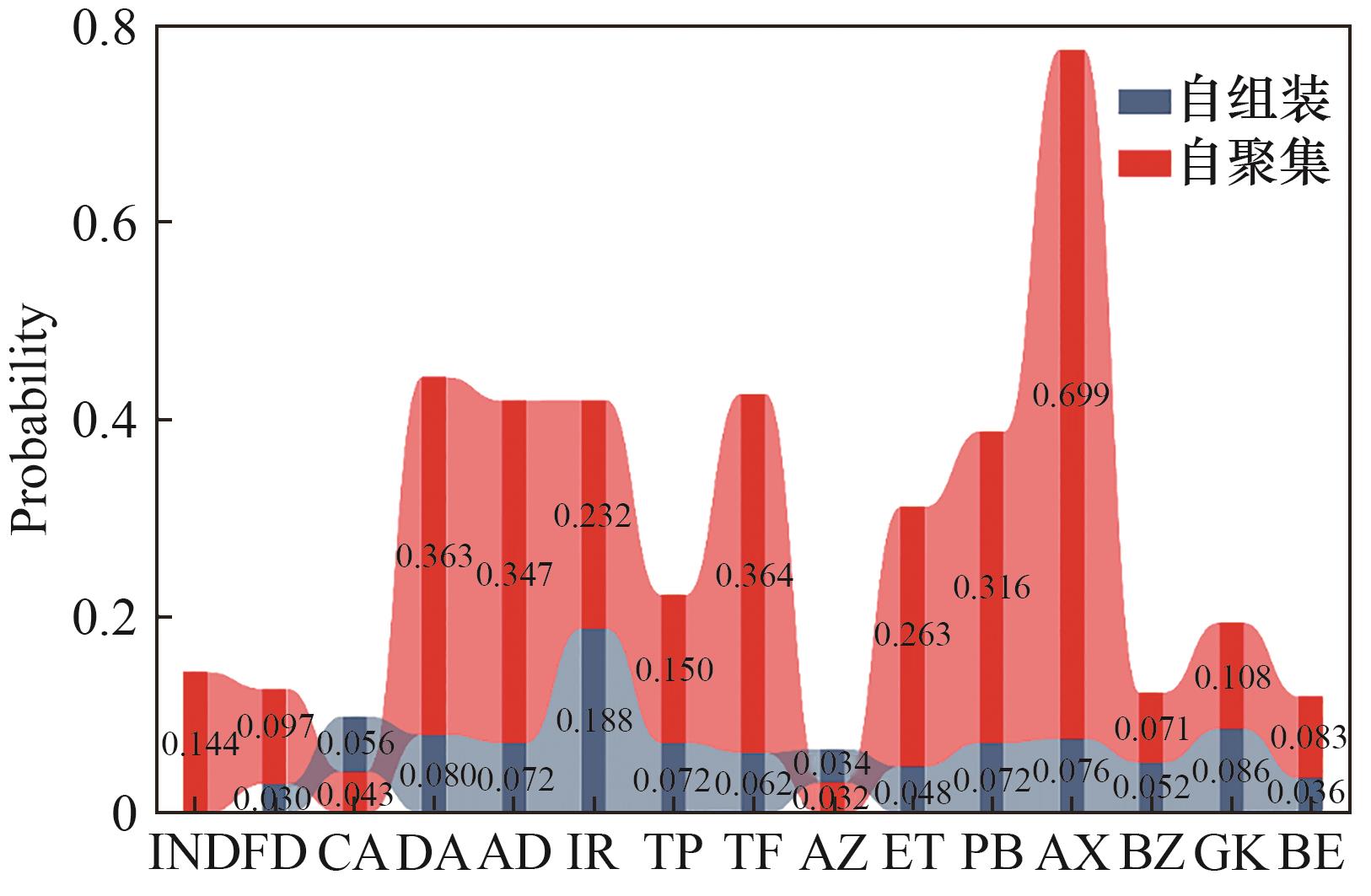
图7 机器学习模型预测药物分子自聚集及与吲哚美辛分子形成自组装的概率
Fig.7 Probability of self-aggregation and self-assembly with indomethacin for the candidate drugs predicted by the ML model
| 1 | Sung H, Ferlay J, Siegel R L, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA: A Cancer Journal for Clinicians, 2021, 71(3): 209-249. |
| 2 | Xiao Y T, Liu J, Guo M Y, et al. Synergistic combination chemotherapy using carrier-free celastrol and doxorubicin nanocrystals for overcoming drug resistance[J]. Nanoscale, 2018, 10(26): 12639-12649. |
| 3 | Salvador-Morales C, Grodzinski P. Nanotechnology tools enabling biological discovery[J]. ACS Nano, 2022, 16(4): 5062-5084. |
| 4 | Liu K F, Liu Y X, Li C X, et al. Self-assembled pH and redox dual responsive carboxymethylcellulose-based polymeric nanoparticles for efficient anticancer drug codelivery[J]. ACS Biomaterials Science & Engineering, 2018, 4(12): 4200-4207. |
| 5 | Kianfar E. Protein nanoparticles in drug delivery: animal protein, plant proteins and protein cages, albumin nanoparticles[J]. Journal of Nanobiotechnology, 2021, 19(1): 159. |
| 6 | Liu Y B, Castro Bravo K M, Liu J W. Targeted liposomal drug delivery: a nanoscience and biophysical perspective[J]. Nanoscale Horizons, 2021, 6(2): 78-94. |
| 7 | Kheraldine H, Rachid O, Habib A M, et al. Emerging innate biological properties of nano-drug delivery systems: a focus on PAMAM dendrimers and their clinical potential[J]. Advanced Drug Delivery Reviews, 2021, 178: 113908. |
| 8 | Wang X W, Zhong X Y, Li J X, et al. Inorganic nanomaterials with rapid clearance for biomedical applications[J]. Chemical Society Reviews, 2021, 50(15): 8669-8742. |
| 9 | Kuang Y T, Li Z K, Chen H, et al. Advances in self-assembled nanotechnology in tumor therapy[J]. Colloids and Surfaces B: Biointerfaces, 2024, 237: 113838. |
| 10 | Zhang X B, Li N, Zhang S W, et al. Emerging carrier-free nanosystems based on molecular self-assembly of pure drugs for cancer therapy[J]. Medicinal Research Reviews, 2020, 40(5): 1754-1775. |
| 11 | 刘雨婷, 王悦全, 张申武, 等. 小分子自组装纳米递药系统研究进展[J]. 药学学报, 2023, 58(3): 516-529. |
| Liu Y T, Wang Y Q, Zhang S W, et al. Advance on small molecule self-assembled nano-drug delivery system[J]. Acta Pharmaceutica Sinica, 2023, 58(3): 516-529. | |
| 12 | Xu X L, Liu A, Liu S Q, et al. Application of molecular dynamics simulation in self-assembled cancer nanomedicine[J]. Biomaterials Research, 2023, 27(1): 39. |
| 13 | Fang F, Chen X Y. Carrier-free nanodrugs: from bench to bedside[J]. ACS Nano, 2024, 18(35): 23827-23841. |
| 14 | Zhong T, Hao Y L, Yao X, et al. Effect of XlogP and Hansen solubility parameters on small molecule modified paclitaxel anticancer drug conjugates self-assembled into nanoparticles[J]. Bioconjugate Chemistry, 2018, 29(2): 437-444. |
| 15 | Reker D, Rybakova Y, Kirtane A R, et al. Computationally guided high-throughput design of self-assembling drug nanoparticles[J]. Nature Nanotechnology, 2021, 16(6): 725-733. |
| 16 | Shamay Y, Shah J, Işık M, et al. Quantitative self-assembly prediction yields targeted nanomedicines[J]. Nature Materials, 2018, 17(4): 361-368. |
| 17 | Frisch M J, Trucks G W, Schlegel H B, et al. Gaussian 16 Rev. A.03 [CP]. Wallingford, CT, 2016. |
| 18 | Zhao Y, Truhlar D G. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: two new functionals and systematic testing of four M06-class functionals and 12 other functionals[J]. Theoretical Chemistry Accounts, 2008, 120(1): 215-241. |
| 19 | Grimme S, Antony J, Ehrlich S, et al. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu[J]. Journal of Chemical Physics, 2010, 132(15): 154104. |
| 20 | Klamt A, Schüürmann G. COSMO: a new approach to dielectric screening in solvents with explicit expressions for the screening energy and its gradient[J]. Journal of the Chemical Society, Perkin Transactions 2, 1993(5): 799-805. |
| 21 | Xantheas S S. On the importance of the fragment relaxation energy terms in the estimation of the basis set superposition error correction to the intermolecular interaction energy[J]. The Journal of Chemical Physics, 1996, 104(21): 8821-8824. |
| 22 | Lu T, Chen F W. Multiwfn: a multifunctional wavefunction analyzer[J]. Journal of Computational Chemistry, 2012, 33(5): 580-592. |
| 23 | Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics[J]. Journal of Molecular Graphics, 1996, 14(1): 33-38. |
| 24 | Lu T, Chen Q. Visualization analysis of weak interactions in chemical systems [M]//YáñEZ M, Boyd R J. Comprehensive Computational Chemistry. Oxford: Elsevier, 2024: 240-264. |
| 25 | Lu T, Chen Q X. Independent gradient model based on hirshfeld partition: a new method for visual study of interactions in chemical systems[J]. Journal of Computational Chemistry, 2022, 43(8): 539-555. |
| 26 | Abraham M J, Murtola T, Schulz R, et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers[J]. SoftwareX, 2015, 1: 19-25. |
| 27 | Wang J M, Wolf R M, Caldwell J W, et al. Development and testing of a general amber force field[J]. Journal of Computational Chemistry, 2004, 25(9): 1157-1174. |
| 28 | Martínez L, Andrade R, Birgin E G, et al. PACKMOL: a package for building initial configurations for molecular dynamics simulations[J]. Journal of Computational Chemistry, 2009, 30(13): 2157-2164. |
| 29 | Berendsen H J C, Postma J P M, van Gunsteren W F, et al. Molecular dynamics with coupling to an external bath[J]. The Journal of Chemical Physics, 1984, 81(8): 3684-3690. |
| 30 | Parrinello M, Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method[J]. Journal of Applied Physics, 1981, 52(12): 7182-7190. |
| 31 | Parrinello M, Rahman A. Strain fluctuations and elastic constants[J]. The Journal of Chemical Physics, 1982, 76(5): 2662-2666. |
| 32 | Hockney R W. The potential calculation and some applications[J]. Methods in Computational Physics, 1970, 9: 136-211. |
| 33 | Darden T, York D, Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems[J]. The Journal of Chemical Physics, 1993, 98(12): 10089-10092. |
| 34 | Essmann U, Perera L, Berkowitz M L, et al. A smooth particle mesh Ewald method[J]. The Journal of Chemical Physics, 1995, 103(19): 8577-8593. |
| 35 | Salem A, Nagy S, Pál S, et al. Reliability of the Hansen solubility parameters as co-crystal formation prediction tool[J]. International Journal of Pharmaceutics, 2019, 558: 319-327. |
| 36 | Shete A, Murthy S, Korpale S, et al. Cocrystals of itraconazole with amino acids: screening, synthesis, solid state characterization, in vitro drug release and antifungal activity[J]. Journal of Drug Delivery Science and Technology, 2015, 28: 46-55. |
| 37 | Mohammad M A, Alhalaweh A, Velaga S P. Hansen solubility parameter as a tool to predict cocrystal formation[J]. International Journal of Pharmaceutics, 2011, 407(1/2): 63-71. |
| 38 | Bagley E, Nelson T, Scigliano J. Three-dimensional solubility parameters and their relationship to internal pressure measurements in polar and hydrogen bonding solvents[J]. Journal of Paint Technology, 1971, 43(555): 35-42. |
| 39 | Breitkreutz J. Prediction of intestinal drug absorption properties by three-dimensional solubility parameters[J]. Pharmaceutical Research, 1998, 15(9): 1370-1375. |
| 40 | Klamt A. Conductor-like screening model for real solvents: a new approach to the quantitative calculation of solvation phenomena[J]. The Journal of Physical Chemistry, 1995, 99(7): 2224-2235. |
| 41 | Loschen C, Klamt A. Solubility prediction, solvate and cocrystal screening as tools for rational crystal engineering[J]. Journal of Pharmacy and Pharmacology, 2015, 67(6): 803-811. |
| 42 | Klamt A, Eckert F, Hornig M, et al. Prediction of aqueous solubility of drugs and pesticides with COSMO-RS[J]. Journal of Computational Chemistry, 2002, 23(2): 275-281. |
| 43 | Eckert F, Klamt A. Fast solvent screening via quantum chemistry: COSMO-RS approach[J]. AIChE Journal, 2002, 48(2): 369-385. |
| 44 | 米泽豪, 花儿. 基于DFT和COSMO-RS理论研究多元胺型离子液体吸收SO2气体[J]. 化工学报, 2023, 74(9): 3681-3696. |
| Mi Z H, Hua E. DFT and COSMO-RS theoretical analysis of SO2 absorption by polyamines type ionic liquids[J]. CIESC Journal, 2023, 74(9): 3681-3696. | |
| 45 | Cao Z X, Wu X J, Wei X H. Ionic liquid screening for desulfurization of coke oven gas based on COSMO-SAC model and process simulation[J]. Chemical Engineering Research and Design, 2021, 176: 146-161. |
| 46 | Mohan M, Keasling J D, Simmons B A, et al. In silico COSMO-RS predictive screening of ionic liquids for the dissolution of plastic[J]. Green Chemistry, 2022, 24(10): 4140-4152. |
| [1] | 王新月, 徐小虎, 张海洋, 尹春华. 维生素A醋酸酯/环糊精包合及性质研究[J]. 化工学报, 2024, 75(S1): 321-328. |
| [2] | 吴哲明, 张碧云, 郑仁朝. 腈水解酶立体选择性改造及其合成布瓦西坦[J]. 化工学报, 2024, 75(7): 2633-2643. |
| [3] | 秦晗淞, 李国梁, 闫昊, 冯翔, 刘熠斌, 陈小博, 杨朝合. 多级孔ZSM-5分子筛中油酸甲酯催化裂解吸附和扩散行为模拟研究[J]. 化工学报, 2024, 75(5): 1870-1881. |
| [4] | 周康, 王建新, 于海, 魏朝良, 范丰奇, 车昕昊, 张磊. 基于分子动力学模拟的矿物基础油泡沫破裂性能研究[J]. 化工学报, 2024, 75(4): 1668-1678. |
| [5] | 刘东飞, 张帆, 刘铮, 卢滇楠. 机器学习势及其在分子模拟中的应用综述[J]. 化工学报, 2024, 75(4): 1241-1255. |
| [6] | 张政, 汪妩琼, 张雅静, 王康军, 吉远辉. 理论计算在药物制剂设计中的研究进展[J]. 化工学报, 2024, 75(4): 1429-1438. |
| [7] | 文一如, 付佳, 刘大欢. 基于机器学习的MOFs材料研究进展:能源气体吸附分离[J]. 化工学报, 2024, 75(4): 1370-1381. |
| [8] | 曾玉娇, 肖炘, 杨刚, 张意博, 郑光明, 李防, 汪凤玲. 基于机理与数据混合驱动的湿法磷酸生产过程代理建模与优化[J]. 化工学报, 2024, 75(3): 936-944. |
| [9] | 陈宇翔, 刘传磊, 龚子君, 赵起越, 郭冠初, 姜豪, 孙辉, 沈本贤. 机器学习辅助乙硫醇高效吸收溶剂分子设计[J]. 化工学报, 2024, 75(3): 914-923. |
| [10] | 吴凡, 彭旭东, 江锦波, 孟祥铠, 梁杨杨. 分子动力学模拟预测天然气密度和黏度的可行性研究[J]. 化工学报, 2024, 75(2): 450-462. |
| [11] | 刘根, 孙仲顺, 张博, 张榕江, 吴志强, 杨伯伦. 机器学习驱动的生物质热解模型建立及挥发分化学链重整制氢工艺优化[J]. 化工学报, 2024, 75(11): 4333-4347. |
| [12] | 付敏, 陈子健, 汤帅, 钱锡亮, 危增曦, 邹昀, 童张法. PVA杂化膜中小分子吸附和扩散行为的分子模拟[J]. 化工学报, 2024, 75(11): 4152-4161. |
| [13] | 阮见, 李双, 温正慧. 自动化与智能化在流动化学中的应用[J]. 化工学报, 2024, 75(11): 4120-4140. |
| [14] | 王茂先, 孙启典, 付哲, 华放, 纪晔, 程易. 分子水平动力学模型和机器学习方法相结合研究废弃塑料热解[J]. 化工学报, 2024, 75(11): 4320-4332. |
| [15] | 汤涵, 蔡进, 覃海航, 陈光进, 孙长宇. 水合物共存体系中气体溶解度预测模型[J]. 化工学报, 2024, 75(11): 4348-4358. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备 11010102001995号
京公网安备 11010102001995号