化工学报 ›› 2019, Vol. 70 ›› Issue (10): 3712-3721.DOI: 10.11949/0438-1157.20190702
谢泽雄1,2( ),陈祥荣1,2,肖文海1,2,李炳志1,2,元英进1,2(
),陈祥荣1,2,肖文海1,2,李炳志1,2,元英进1,2( )
)
收稿日期:2019-06-21
修回日期:2019-08-31
出版日期:2019-10-05
发布日期:2019-10-05
通讯作者:
元英进
作者简介:谢泽雄(1990—),男,博士,副教授,基金资助:
Zexiong XIE1,2( ),Xiangrong CHEN1,2,Wenhai XIAO1,2,Bingzhi LI1,2,Yingjin YUAN1,2(
),Xiangrong CHEN1,2,Wenhai XIAO1,2,Bingzhi LI1,2,Yingjin YUAN1,2( )
)
Received:2019-06-21
Revised:2019-08-31
Online:2019-10-05
Published:2019-10-05
Contact:
Yingjin YUAN
摘要:
细胞工厂能利用微生物细胞制备人类所需能源、药物和化学品。底盘细胞和外源代谢路径的适配是构建高效细胞工厂的核心难题。基因组再造指利用化学合成的核苷酸分子“自下而上”构建生物基因组,基因组诱导重排指通过在全基因组尺度进行DNA序列与结构的人为调控。基因组再造和诱导重排实现了对生命体的创造,增强了模式底盘细胞的遗传稳定性和操作柔性。基因组的适度精简和密码子简化改善细胞对底物、能量的利用效率,提高细胞生理性能的预测性和可控性。基因组重排可诱导染色体发生随机删除、复制、移位和倒置等结构变异,可产生大量性状优良的模式底盘细胞,进而加速代谢路径优化,提高路径和底盘细胞的适配性,为人工细胞工厂快速构建和优化提供了新策略。
中图分类号:
谢泽雄, 陈祥荣, 肖文海, 李炳志, 元英进. 基因组再造与重排构建细胞工厂[J]. 化工学报, 2019, 70(10): 3712-3721.
Zexiong XIE, Xiangrong CHEN, Wenhai XIAO, Bingzhi LI, Yingjin YUAN. Cell factory construction accelerated by genome synthesis and rearrangement[J]. CIESC Journal, 2019, 70(10): 3712-3721.
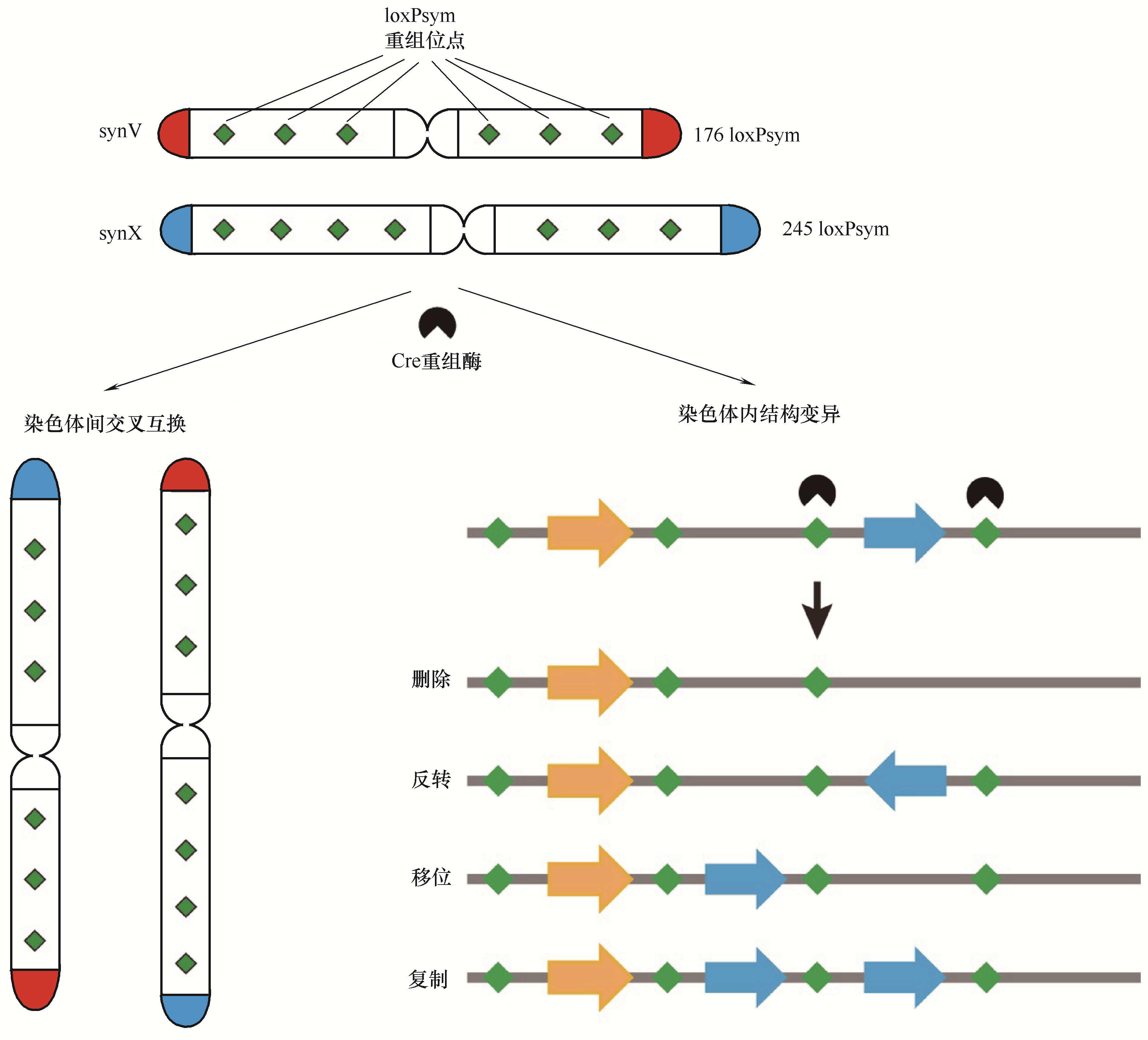
图3 合成型酿酒酵母SCRaMbLE系统加速基因组重排,产生多样性基因组结构变异
Fig.3 SCRaMbLE system of synthetic yeast accelerates genome rearrangement, generating diverse genomic structural variations
| 1 | Breitling R , Takano E . Synthetic biology advances for pharmaceutical production[J]. Curr. Opin. Biotechnol., 2015, 35: 46-51. |
| 2 | Chang M C , Eachus R A , Trieu W , et al . Engineering Escherichia coli for production of functionalized terpenoids using plant P450s[J]. Nat. Chem. Biol., 2007, 3(5): 274-277. |
| 3 | Paddon C J , Keasling J D . Semi-synthetic artemisinin: a model for the use of synthetic biology in pharmaceutical development[J]. Nat. Rev. Microbiol., 2014, 12(5): 355-367. |
| 4 | Hong K K , Nielsen J . Metabolic engineering of Saccharomyces cerevisiae: a key cell factory platform for future biorefineries[J]. Cell Mol. Life Sci., 2012, 69(16): 2671-2690. |
| 5 | Ostrov N , Landon M , Guell M , et al . Design, synthesis, and testing toward a 57-codon genome[J]. Science, 2016, 353(6301): 819-822. |
| 6 | Fredens J , Wang K , de la Torre D , et al . Total synthesis of Escherichia coli with a recoded genome[J]. Nature, 2019, 569(7757): 514-518. |
| 7 | Pennisi E . Building the ultimate yeast genome[J]. Science, 2014, 343(6178): 1426-1429. |
| 8 | Sliva A , Yang H , Boeke J D , et al . Freedom and responsibility in synthetic genomics: the synthetic yeast project[J]. Genetics, 2015, 200(4): 1021-1028. |
| 9 | Dymond J S , Richardson S M , Coombes C E , et al . Synthetic chromosome arms function in yeast and generate phenotypic diversity by design[J]. Nature, 2011, 477(7365): 471-476. |
| 10 | Annaluru N , Muller H , Mitchell L A , et al . Total synthesis of a functional designer eukaryotic chromosome[J]. Science, 2014, 344(6179): 55-58. |
| 11 | Oliver S G , van der Aart Q J , Agostoni-Carbone M L , et al . The complete DNA sequence of yeast chromosome III[J]. Nature, 1992, 357(6373): 38-46. |
| 12 | Mercy G , Mozziconacci J , Scolari V F , et al . 3D organization of synthetic and scrambled chromosomes[J]. Science, 2017, 355(6329): eaaf4597 |
| 13 | Mitchell L A , Wang A , Stracquadanio G , et al . Synthesis, debugging, and effects of synthetic chromosome consolidation: synVI and beyond[J]. Science, 2017, 355(6329): eaaf4831. |
| 14 | Richardson S M , Mitchell L A , Stracquadanio G , et al . Design of a synthetic yeast genome[J]. Science, 2017, 355(6329): 1040-1044. |
| 15 | Shen Y , Wang Y , Chen T , et al . Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome[J]. Science, 2017, 355(6329): eaaf4791. |
| 16 | Wu Y , Li B Z , Zhao M , et al . Bug mapping and fitness testing of chemically synthesized chromosome X[J]. Science, 2017, 355(6329): eaaf4706. |
| 17 | Xie Z X , Li B Z , Mitchell L A , et al . “Perfect” designer chromosome V and behavior of a ring derivative[J]. Science, 2017, 355(6329): eaaf4704. |
| 18 | Xie Z X , Liu D , Li B Z , et al . Design and chemical synthesis of eukaryotic chromosomes[J]. Chem. Soc. Rev., 2017, 46(23): 7191-7207. |
| 19 | Zhang W , Zhao G , Luo Z , et al . Engineering the ribosomal DNA in a megabase synthetic chromosome[J]. Science, 2017, 355(6329): eaaf3981. |
| 20 | Gibson D G , Glass J I , Lartigue C , et al . Creation of a bacterial cell controlled by a chemically synthesized genome[J]. Science, 2010, 329(5987): 52-56. |
| 21 | C A 3rd Hutchison , Chuang R Y , Noskov V N , et al . Design and synthesis of a minimal bacterial genome[J]. Science, 2016, 351(6280): aad6253. |
| 22 | Goffeau A , Barrell B G , Bussey H , et al . Life with 6000 genes[J]. Science, 1996, 274(5287): 546-547, 563. |
| 23 | Giaever G , Chu A M , Ni L , et al . Functional profiling of the Saccharomyces cerevisiae genome[J]. Nature, 2002, 418(6896): 387-391. |
| 24 | Tong A H , Evangelista M , Parsons A B , et al . Systematic genetic analysis with ordered arrays of yeast deletion mutants[J]. Science, 2001, 294(5550): 2364-2368. |
| 25 | Tong A H , Lesage G , Bader G D , et al . Global mapping of the yeast genetic interaction network[J]. Science, 2004, 303(5659): 808-813. |
| 26 | Costanzo M , Baryshnikova A , Bellay J , et al . The genetic landscape of a cell[J]. Science, 2010, 327(5964): 425-431. |
| 27 | Costanzo M , VanderSluis B , Koch E N , et al . A global genetic interaction network maps a wiring diagram of cellular function[J]. Science, 2016, 353(6306): aaf1420 |
| 28 | Cello J , Paul A V , Wimmer E . Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template[J]. Science, 2002, 297(5583): 1016-1018. |
| 29 | Smith H O , Hutchison C A , Pfannkoch C , et al . Generating a synthetic genome by whole genome assembly: X174 bacteriophage from synthetic oligonucleotides[J]. Proc. Natl. Acad. Sci. USA, 2003, 100(26): 15440-15445. |
| 30 | Chan L Y , Kosuri S , Endy D . Refactoring bacteriophage T7[J]. Mol. Syst. Biol., 2005, 1(1): 2005.0018. |
| 31 | Becker M M , Graham R L , Donaldson E F , et al . Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice[J]. Proc. Natl. Acad. Sci. USA, 2008, 105(50): 19944-19949. |
| 32 | Orlinger K K , Holzer G W , Schwaiger J , et al . An inactivated West Nile Virus vaccine derived from a chemically synthesized cDNA system[J]. Vaccine, 2010, 28(19): 3318-3324. |
| 33 | Itaya M , Fujita K , Kuroki A , et al . Bottom-up genome assembly using the Bacillus subtilis genome vector[J]. Nat. Methods, 2008, 5(1): 41-43. |
| 34 | Gibson D G , Smith H O , Hutchison C A , et al . Chemical synthesis of the mouse mitochondrial genome[J]. Nat. Methods, 2010, 7(11): 901-903. |
| 35 | Lajoie M J , Rovner A J , Goodman D B , et al . Genomically recoded organisms expand biological functions[J]. Science, 2013, 342(6156): 357-360. |
| 36 | Liu W , Luo Z , Wang Y , et al . Rapid pathway prototyping and engineering using in vitro and in vivo synthetic genome SCRaMbLE-in methods[J]. Nat. Commun., 2018, 9(1): 1936. |
| 37 | Dymond J , Boeke J . The Saccharomyces cerevisiae SCRaMbLE system and genome minimization[J]. Bioeng. Bugs., 2012, 3(3): 168-171. |
| 38 | Hoess R H , Wierzbicki A , Abremski K . The role of the loxP spacer region in P1 site-specific recombination[J]. Nucleic. Acids. Res., 1986, 14(5): 2287-2300. |
| 39 | Testa G , Stewart A F . Creating a transloxation[J]. Embo. Reports, 2000, 1(2): 120-121. |
| 40 | Glass J I , Assad-Garcia N , Alperovich N , et al . Essential genes of a minimal bacterium[J]. Proc. Natl. Acad. Sci. USA, 2006, 103(2): 425-430. |
| 41 | Lajoie M J , Kosuri S , Mosberg J A , et al . Probing the limits of genetic recoding in essential genes[J]. Science, 2013, 342(6156): 361-363. |
| 42 | Neumann H , Hancock S M , Buning R , et al . A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation[J]. Mol. Cell, 2009, 36(1): 153-163. |
| 43 | Park H S , Hohn M J , Umehara T , et al . Expanding the genetic code of Escherichia coli with phosphoserine[J]. Science, 2011, 333(6046): 1151-1154. |
| 44 | Xie J , Schultz P G . A chemical toolkit for proteins—an expanded genetic code[J]. Nat. Rev. Mol. Cell Biol., 2006, 7(10): 775-782. |
| 45 | Wang L , Xie J , Schultz P G . Expanding the genetic code[J]. Annu. Rev. Biophys. Biomol. Struct., 2006, 35: 225-249. |
| 46 | Maxmen A . Synthetic yeast chromosomes help probe mysteries of evolution[J]. Nature, 2017, 543(7645): 298-299. |
| 47 | Isaacs F J , Carr P A , Wang H H , et al . Precise manipulation of chromosomes in vivo enables genome-wide codon replacement[J]. Science, 2011, 333(6040): 348-353. |
| 48 | Wang H H , Isaacs F J , Carr P A , et al . Programming cells by multiplex genome engineering and accelerated evolution[J]. Nature, 2009, 460(7257): 894-898. |
| 49 | Mandell D J , Lajoie M J , Mee M T , et al . Biocontainment of genetically modified organisms by synthetic protein design[J]. Nature, 2015, 518(7537): 55-60. |
| 50 | Rovner A J , Haimovich A D , Katz S R , et al . Recoded organisms engineered to depend on synthetic amino acids[J]. Nature, 2015, 518(7537): 89-93. |
| 51 | Ma N J , Moonan D W , Isaacs F J . Precise manipulation of bacterial chromosomes by conjugative assembly genome engineering[J]. Nat. Protoc., 2014, 9(10): 2285-2300. |
| 52 | Wang K , Fredens J , Brunner S F , et al . Defining synonymous codon compression schemes by genome recoding[J]. Nature, 2016, 539(7627): 59-64. |
| 53 | Salim H M , Ring K L , Cavalcanti A R . Patterns of codon usage in two ciliates that reassign the genetic code: Tetrahymena thermophila and Paramecium tetraurelia [J]. Protist, 2008, 159(2): 283-298. |
| 54 | Gorter de Vries A R , Pronk J T , Daran J G . Industrial relevance of chromosomal copy number variation in Saccharomyces yeasts[J]. Appl. Environ. Microbiol., 2017, 83(11): e03206-16. |
| 55 | Takagi A , Harashima S , Oshima Y . Construction and characterization of isogenic series of Saccharomyces cerevisiae polyploid strains[J]. Appl. Environ. Microbiol., 1983, 45(3): 1034-1040. |
| 56 | Li Y , Wu Y , Ma L , et al . Loss of heterozygosity by SCRaMbLEing[J]. Sci. China Life Sci., 2019, 62(3): 381-393. |
| 57 | Shao Y , Lu N , Wu Z , et al . Creating a functional single-chromosome yeast[J]. Nature, 2018, 560(7718): 331-335. |
| 58 | Luo J , Sun X , Cormack B P , et al . Karyotype engineering by chromosome fusion leads to reproductive isolation in yeast[J]. Nature, 2018, 560(7718): 392-396. |
| 59 | Neurohr G , Naegeli A , Titos I , et al . A midzone-based ruler adjusts chromosome compaction to anaphase spindle length[J]. Science, 2011, 332(6028): 465-468. |
| 60 | Wang J , Xie Z X , Ma Y , et al . Ring synthetic chromosome V SCRaMbLE[J]. Nat. Commun., 2018, 9(1): 3783. |
| 61 | Weischenfeldt J , Symmons O , Spitz F , et al . Phenotypic impact of genomic structural variation: insights from and for human disease[J]. Nat. Rev. Genet., 2013, 14(2): 125-138. |
| 62 | Blount B A , Gowers G F , Ho J C H , et al . Rapid host strain improvement by in vivo rearrangement of a synthetic yeast chromosome[J]. Nat. Commun., 2018, 9(1): 1932. |
| 63 | Jia B , Wu Y , Li B Z , et al . Precise control of SCRaMbLE in synthetic haploid and diploid yeast[J]. Nat. Commun., 2018, 9(1): 1933. |
| 64 | Jullesson D , David F , Pfleger B , et al . Impact of synthetic biology and metabolic engineering on industrial production of fine chemicals[J]. Biotechnol. Adv., 2015, 33(7): 1395-1402. |
| 65 | Luo Z , Wang L , Wang Y , et al . Identifying and characterizing SCRaMbLEd synthetic yeast using ReSCuES[J]. Nat. Commun., 2018, 9(1): 1930. |
| 66 | Ma L , Li Y , Chen X , et al . SCRaMbLE generates evolved yeasts with increased alkali tolerance[J]. Microb. Cell Fact., 2019, 18(1): 52. |
| 67 | Shen M J , Wu Y , Yang K , et al . Heterozygous diploid and interspecies SCRaMbLEing[J]. Nat. Commun., 2018, 9(1): 1934. |
| 68 | Fisher A K , Freedman B G , Bevan D R , et al . A review of metabolic and enzymatic engineering strategies for designing and optimizing performance of microbial cell factories[J]. Comput. Struct. Biotechnol. J., 2014, 11(18): 91-99. |
| 69 | Wu Y , Zhu R Y , Mitchell L A , et al . In vitro DNA SCRaMbLE[J]. Nat. Commun., 2018, 9(1): 1935. |
| 70 | Hochrein L , Mitchell L A , Schulz K , et al . L-SCRaMbLE as a tool for light-controlled cre-mediated recombination in yeast[J]. Nat. Commun., 2018, 9(1): 1931. |
| [1] | 孙怡, 张腾, 吕波, 李春. 胞内生物传感器提高微生物细胞工厂的精细调控[J]. 化工学报, 2022, 73(2): 521-534. |
| [2] | 王欣慧, 王颖, 姚明东, 肖文海. 维生素A生物合成的研究进展[J]. 化工学报, 2022, 73(10): 4311-4323. |
| [3] | 郑煜堃, 孙青, 陈振, 于慧敏. 微生物细胞工厂生产化学品的研究进展——以几种典型小分子和大分子化学品为例[J]. 化工学报, 2021, 72(12): 6109-6121. |
| [4] | 张震, 曾雪城, 秦磊, 李春. 微生物细胞工厂的智能设计进展[J]. 化工学报, 2021, 72(12): 6093-6108. |
| [5] | 王凯峰, 王金鹏, 韦萍, 纪晓俊. 代谢工程改造解脂耶氏酵母生产脂肪酸及其衍生物[J]. 化工学报, 2021, 72(1): 351-365. |
| [6] | 薛海洁, 王颖, 李春. 植物天然产物的微生物合成与转化[J]. 化工学报, 2019, 70(10): 3825-3835. |
| [7] | 姚善泾, 蔡礼年, 林东强. 黑曲霉作为分泌蛋白细胞工厂的研究进展[J]. 化工学报, 2019, 70(10): 3690-3703. |
| [8] | 肖文海, 王颖, 元英进. 化学品绿色制造核心技术——合成生物学[J]. 化工学报, 2016, 67(1): 119-128. |
| [9] | 赵雨佳, 张根林, 周晓宏, 李春. 核糖核酸开关用于微生物细胞工厂的智能与精细调控[J]. 化工学报, 2015, 66(10): 3811-2819. |
| [10] | 夏燕春, 王超, 陈园, 吴江磊, 邹球龙, 张晓琳, 李春. 基于基因组重排技术的多杀菌素高产菌株选育[J]. 化工学报, 2014, 65(9): 0-0. |
| [11] | 夏燕春, 王超, 陈园, 吴江磊, 邹球龙, 张晓琳, 李春. 基于基因组重排技术的多杀菌素高产菌株选育[J]. 化工学报, 2014, 65(9): 3576-3582. |
| [12] | 李寅;曹竹安. 微生物代谢工程:绘制细胞工厂的蓝图 [J]. CIESC Journal, 2004, 55(10): 1573-1580. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备 11010102001995号
京公网安备 11010102001995号