化工学报 ›› 2020, Vol. 71 ›› Issue (9): 4058-4070.DOI: 10.11949/0438-1157.20200460
收稿日期:2020-04-30
修回日期:2020-05-21
出版日期:2020-09-05
发布日期:2020-09-05
通讯作者:
王佳,袁其朋
作者简介:高虎涛(1997—),男,硕士研究生,基金资助:
Hutao GAO( ),Xiaolin SHEN,Xinxiao SUN,Jia WANG(
),Xiaolin SHEN,Xinxiao SUN,Jia WANG( ),Qipeng YUAN(
),Qipeng YUAN( )
)
Received:2020-04-30
Revised:2020-05-21
Online:2020-09-05
Published:2020-09-05
Contact:
Jia WANG,Qipeng YUAN
摘要:
氨基酸的工业化生产已有上百年历史,多用于动物饲料和食品添加剂;很多种类的氨基酸如L-半胱氨酸、β-丙氨酸、S-腺苷甲硫氨酸、4-羟基异亮氨酸和高丝氨酸等也具有很高的应用价值。相较于化工合成或分离提取的方式,利用微生物细胞作为平台生产氨基酸及其衍生物具有绿色安全、可持续等独特的优势。本文综述了近年来微生物合成氨基酸及其衍生物的研究进展,分别介绍了碳源的高效利用、限速步骤的调节、碳通量的调节、转录和反馈抑制调节以及转运调节等代谢调控策略在提高微生物生产氨基酸及其衍生物效率的研究及应用,分析了不同调控策略的优势和缺点,总结了不同氨基酸及其衍生物的应用价值,最后展望了微生物作为细胞工厂生产各类氨基酸及其衍生物的广阔前景。
中图分类号:
高虎涛, 申晓林, 孙新晓, 王佳, 袁其朋. 代谢工程调控策略在生物合成氨基酸及其衍生物中的应用[J]. 化工学报, 2020, 71(9): 4058-4070.
Hutao GAO, Xiaolin SHEN, Xinxiao SUN, Jia WANG, Qipeng YUAN. Metabolic engineering strategies in biosynthesis of amino acids and their derivatives[J]. CIESC Journal, 2020, 71(9): 4058-4070.

图1 碳源高效利用策略的应用(a)L-赖氨酸在谷氨酸棒状杆菌中的部分生物合成途径;(b)L-3,4-二羟基苯丙氨酸在大肠杆菌中的生物合成途径;(c)S-腺苷甲硫氨酸在解淀粉芽孢杆菌中的生物合成途径;(d)4-羟基异亮氨酸在大肠杆菌中的生物合成途径IolT1, IolT2—肌醇通透酶;EIIbglF—β-葡萄糖苷-PTS渗透酶;GlK,PpgK,Cgl2647—葡萄糖激酶; IICBGlc—整合膜葡萄糖通透酶;galP—半乳糖渗透酶基因; glk—葡萄糖激酶基因; sucC—琥珀酰-CoA合成酶基因;metA—高丝氨酸O-琥珀酰转移酶基因;metB—胱硫醚-γ-半胱氨酸酶基因; IDH—异柠檬酸脱氢酶;IDO—L-异亮氨酸双加氧酶;AceA—异柠檬酸连接酶;AceK—异柠檬酸脱氢酶激酶;SucAB—α-酮戊二酸脱氢酶
Fig.1 Application of carbon source efficient utilization strategy(a) partial biosynthetic pathway of L-lysine in Corynebacterium glutamicum; (b) biosynthetic pathway of L-3,4-dihydroxyphenylalanine in E. coli;(c) biosynthetic pathway of S-adenosylmethionine in B. amyloliquefaciens; (d) biosynthetic pathway of 4-hydroxyisoleucine in E. coli
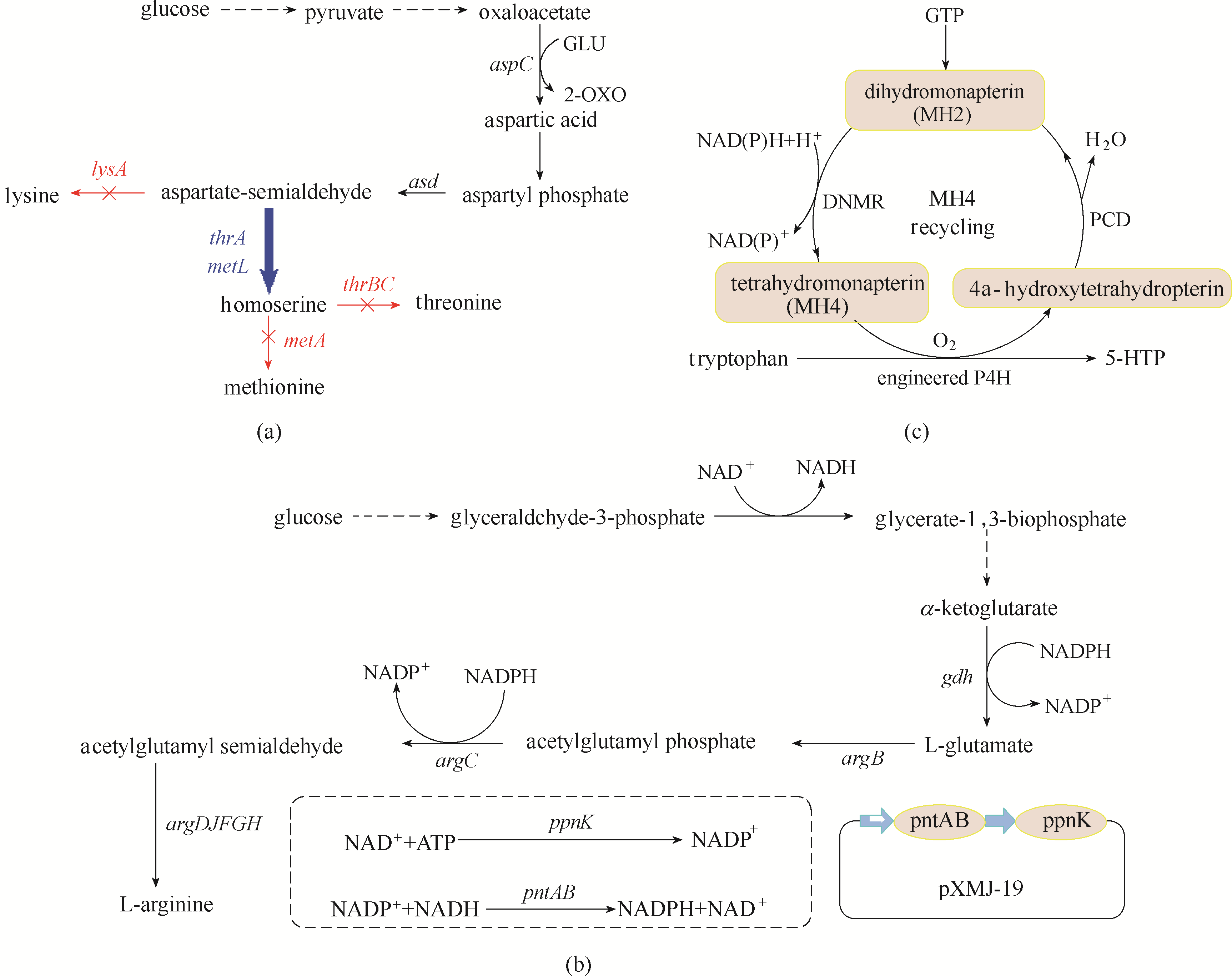
图2 限速步骤调节策略的应用(a)高丝氨酸在大肠杆菌中的生产代谢途径;(b)L-精氨酸在谷氨酸棒状杆菌中的生产代谢途径;(c)5-羟基色氨酸在大肠杆菌中的生产代谢途径aspC—天冬氨酸转氨酶基因;asd—天门冬氨酸半醛脱氢酶基因;lysA—二氨基庚二酸酯脱羧酶基因;thrA—高丝氨酸脱氢酶I基因;metL—高丝氨酸脱氢酶II基因;thrBC—高丝氨酸激酶基因;metA—高丝氨酸琥珀酰转移酶基因;gdh—谷氨酸脱氢酶基因;argB—N-乙酰谷氨酸激酶基因;argC—N-乙酰-γ-谷氨酰磷酸还原酶基因;argD—乙酰鸟氨酸氨基转移酶基因;argJ—双功能鸟氨酸乙酰转移酶基因;argF—鸟氨酸氨基甲酰基转移酶基因;argG—精氨酸琥珀酸合酶基因;argH—精氨酸琥珀酸合酶基因;pntAB—膜结合转氢酶基因;ppnK—NAD+激酶基因;DNMR—二氢莫那还原酶;P4H—苯丙氨酸4-羟化酶;PCD—蝶呤4a-甲醇胺脱水酶
Fig.2 Application of speed-limiting step adjustment strategy(a) the metabolic pathway of homoserine production in E. coli; (b) the metabolic pathway of L-arginine in C. glutamicum; (c) the metabolic pathway of 5-hydroxytryptophan in E. coli

图3 碳通量调节策略的应用(a)L-鸟氨酸在谷氨酸棒状杆菌中的生产代谢途径;(b)β-鸟氨酸在大肠杆菌中的生产代谢途径odhA—酮戊二酸脱氢酶基因; argCJBD—精氨酸合成的操纵子基因;argF—鸟氨酸氨基甲酰基转移酶基因;标记紫色为下调基因,绿色为过表达基因,红色为被敲除基因;ldhA—D-乳酸脱氢酶基因;pflB—丙酮酸甲酸酯裂解酶基因;adhE—酒精/醛脱氢酶基因;pta—磷酸乙酰基转移酶基因;gluDH—谷氨酸脱氢酶;akI,akII和akIII—天冬氨酸激酶基因;BtADC—L-天冬氨酸α-脱羧酶;AspA—天冬氨酸酶;标记蓝色箭头表示基因过表达,红色箭头表示基因敲除
Fig.3 Application of carbon flux regulation strategy(a) the metabolic pathway of L-ornithine in C. glutamicum; (b) production and metabolism pathway of β-ornithine in E. coli
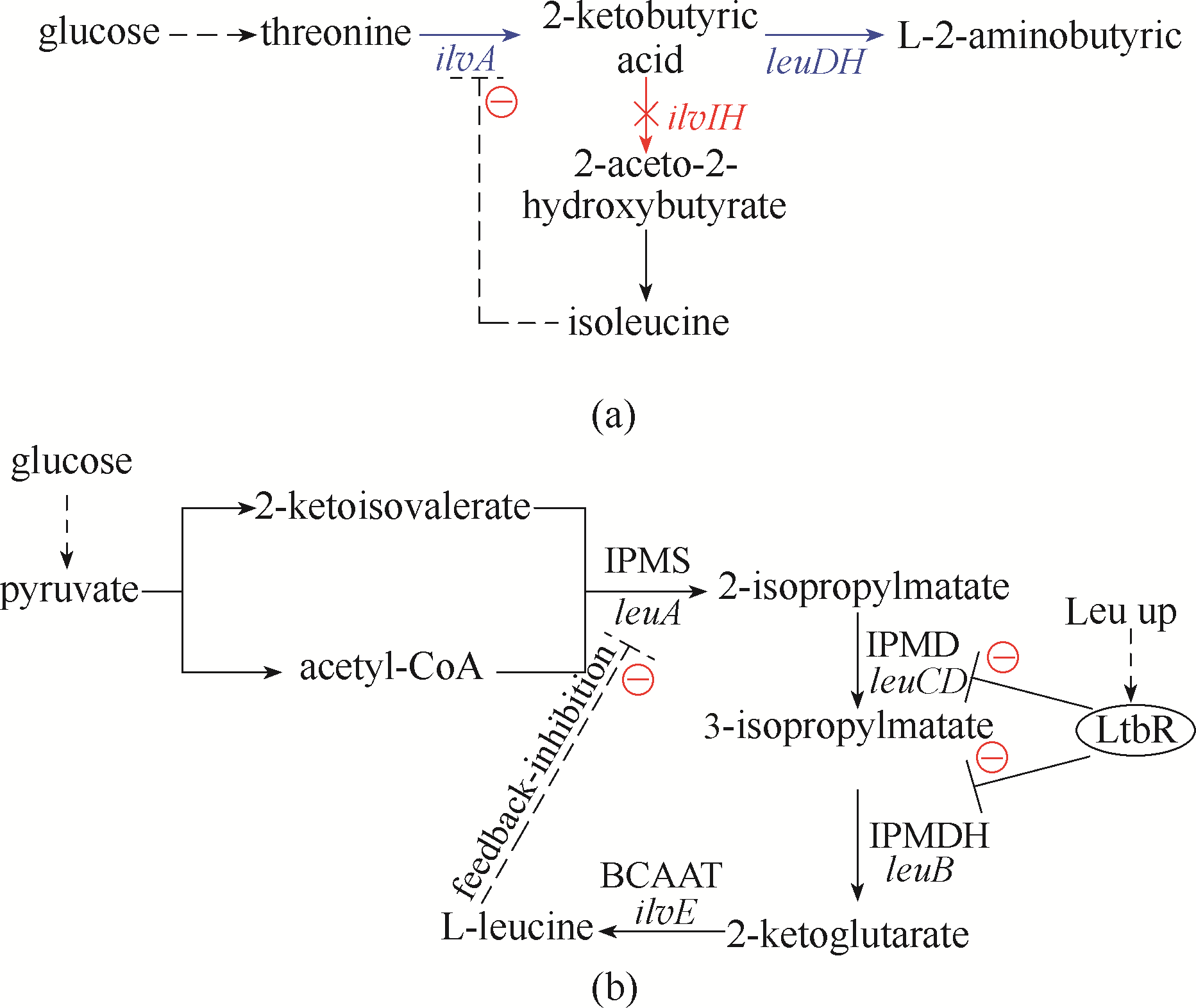
图4 转录调节与反馈抑制调节策略的应用(a)2-氨基丁酸在大肠杆菌中的生产代谢途径;(b)L-亮氨酸在谷氨酸棒状杆菌中的生产代谢途径ilvA—苏氨酸脱水酶基因;ilvIH—乙酰羟酸合酶Ⅲ基因;leuDH—2-酮丁酸脱氢酶基因;减号表示受到抑制,虚线表示反馈抑制,标记蓝色箭头表示过表达;leuA—2-异丙基苹果酸合酶(IPMS)基因;leuCD—3-异丙基苹果酸脱水酶(IPMD)基因;leuB—3-异丙基苹果酸脱氢酶(IPMDH)基因;ilvE—支链氨基酸转氨酶(BCAAT)基因
Fig.4 Application of transcription regulation and feedback inhibition regulation strategies(a) the metabolic pathway of 2-aminobutyric acid in E. coli; (b) L-leucine production and metabolic pathways in C. glutamicum
| 代谢工程策略 | 氨基酸及其衍生物 | 功能 | 工程菌 | 产量 | 文献 |
|---|---|---|---|---|---|
| 碳源的高效利用 | 左旋鸟氨酸 | 一种非蛋白质氨基酸,能够加速伤口愈合并能有效维持心脏健康 | 谷氨酸棒状杆菌S9114 | 43.6 g/L | [ |
| L-亮氨酸 | 在刺激肌肉蛋白质合成和葡萄糖稳态中具有重要作用 | 谷氨酸棒状杆菌 | 23.7 g/L | [ | |
| L-赖氨酸 | 促进人体的发育、增强免疫力,并有提高中枢神经功能的作用 | 谷氨酸棒状杆菌 ZL-19 | 201.6 g/L | [ | |
| L-3,4-二羟基苯丙氨酸 | 用于治疗帕金森氏疾病类药物的主要前体 | 大肠杆菌 | 1.51 g/L | [ | |
| 苯丙氨酸 | 用于生产人造甜味剂阿斯巴甜 | 大肠杆菌 | 1.4 g/L | [ | |
| S-腺苷甲硫氨酸 | 被用作预防和治疗骨关节炎的功能性营养品或药物 | 解淀粉芽孢杆菌 | 107.47mg/L | [ | |
| 4-羟基异亮氨酸 | 在治疗和预防2-型糖尿病中有广阔应用前景 | 大肠杆菌 | 24.1 g/L | [ | |
| 限速步骤的调节 | 高丝氨酸 | 诱导免疫系统,增强植物对疾病的抵抗力 | 大肠杆菌W3110 | 1.20 g/L | [ |
| L-高丙氨酸 | 抗癫痫药物左乙拉西坦和溴拉西坦的直接前体 | 大肠杆菌 | 5.4 g/L | [ | |
| L-精氨酸 | 具有舒张因子的作用,临床上用于舒张和扩张血管 | 谷氨酸棒状杆菌 | 61.13 g/L | [ | |
| L-赖氨酸 | 促进人体的发育、增强免疫力,并有提高中枢神经功能的作用 | 谷氨酸棒状杆菌 | 8.76 g/L | [ | |
| 5-羟基色氨酸 | 用于治疗和缓解抑郁症,有效治疗失眠和慢性头痛 | 大肠杆菌 | 1.11 g/L | [ | |
| 碳通量的调节 | L-鸟氨酸 | 用于治疗肝脏疾病和创伤,在肝脏保护中起着有效的作用 | 谷氨酸棒状杆菌 | 16 g/L | [ |
| 左旋肉碱 | 用于治疗功能障碍疾病 | 大肠杆菌 | 61.3 g/L | [ | |
| β-丙氨酸 | 生物体中泛酸生产的主要前体 | 大肠杆菌 | 43.12 g/L | [ | |
| 5-氨基戊酸酯 | 生产尼龙的结构单元 | 谷氨酸棒状杆菌 | 28 g/L | [ | |
| 5-氨基乙酰丙酸 | 用于多种癌症的光动力学诊断和治疗中 | 谷氨酸棒状杆菌 | 2.06 g/L | [ | |
| 转录调节与反馈抑制调节 | L-酪氨酸 | 食品和饲料添加剂 | 酿酒酵母 | 36.2 mg/L | [ |
| 2-氨基丁酸 | 抗结核性乙胺丁醇的重要前体 | 大肠杆菌 | 5.39 g/L | [ | |
| L-精氨酸 | 在食品和补充保健食品,制药和化妆品行业中有重要应用 | 谷氨酸棒状杆菌 | 61.9 g/L | [ | |
| L-亮氨酸 | 在刺激肌肉蛋白质合成和葡萄糖稳态中具有重要作用 | 谷氨酸棒状杆菌 | 5.7 g/L | [ | |
| L-苏氨酸 | 广泛用于食品、制药和化妆品行业 | 大肠杆菌 | 26.0 g/L | [ | |
| 转运系统的调节 | L-异亮氨酸 | 形成血红蛋白必需氨基酸,在调节血糖与能量方面起到关键作用 | 谷氨酸棒状杆菌 | 26.8 g/L | [ |
| 高丝氨酸 | 诱导免疫系统,增强植物对疾病的抵抗力 | 大肠杆菌W3110 | 39.54 g/L | [ | |
| L-半胱氨酸 | 在提高动物的免疫调节和促进动物生长发育方面起着重要作用 | 大肠杆菌 | 634.4 mg/L | [ | |
| L-蛋氨酸 | 用于脂肪肝,以及酒精等引起的肝损害 | 大肠杆菌 | 593.5 mg/L | [ | |
| L-瓜氨酸 | 对于保持肌肉和肝脏健康有重要作用 | 谷氨酸棒状杆菌 | 21 g/L | [ | |
| L-苏氨酸 | 促进人体发育和抗脂肪肝的药用效能 | 谷氨酸棒状杆菌 | 8.1 g/L | [ |
表1 本文涉及到的氨基酸及其衍生物摘要
Table 1 Summary of amino acids and their derivatives involved in this article
| 代谢工程策略 | 氨基酸及其衍生物 | 功能 | 工程菌 | 产量 | 文献 |
|---|---|---|---|---|---|
| 碳源的高效利用 | 左旋鸟氨酸 | 一种非蛋白质氨基酸,能够加速伤口愈合并能有效维持心脏健康 | 谷氨酸棒状杆菌S9114 | 43.6 g/L | [ |
| L-亮氨酸 | 在刺激肌肉蛋白质合成和葡萄糖稳态中具有重要作用 | 谷氨酸棒状杆菌 | 23.7 g/L | [ | |
| L-赖氨酸 | 促进人体的发育、增强免疫力,并有提高中枢神经功能的作用 | 谷氨酸棒状杆菌 ZL-19 | 201.6 g/L | [ | |
| L-3,4-二羟基苯丙氨酸 | 用于治疗帕金森氏疾病类药物的主要前体 | 大肠杆菌 | 1.51 g/L | [ | |
| 苯丙氨酸 | 用于生产人造甜味剂阿斯巴甜 | 大肠杆菌 | 1.4 g/L | [ | |
| S-腺苷甲硫氨酸 | 被用作预防和治疗骨关节炎的功能性营养品或药物 | 解淀粉芽孢杆菌 | 107.47mg/L | [ | |
| 4-羟基异亮氨酸 | 在治疗和预防2-型糖尿病中有广阔应用前景 | 大肠杆菌 | 24.1 g/L | [ | |
| 限速步骤的调节 | 高丝氨酸 | 诱导免疫系统,增强植物对疾病的抵抗力 | 大肠杆菌W3110 | 1.20 g/L | [ |
| L-高丙氨酸 | 抗癫痫药物左乙拉西坦和溴拉西坦的直接前体 | 大肠杆菌 | 5.4 g/L | [ | |
| L-精氨酸 | 具有舒张因子的作用,临床上用于舒张和扩张血管 | 谷氨酸棒状杆菌 | 61.13 g/L | [ | |
| L-赖氨酸 | 促进人体的发育、增强免疫力,并有提高中枢神经功能的作用 | 谷氨酸棒状杆菌 | 8.76 g/L | [ | |
| 5-羟基色氨酸 | 用于治疗和缓解抑郁症,有效治疗失眠和慢性头痛 | 大肠杆菌 | 1.11 g/L | [ | |
| 碳通量的调节 | L-鸟氨酸 | 用于治疗肝脏疾病和创伤,在肝脏保护中起着有效的作用 | 谷氨酸棒状杆菌 | 16 g/L | [ |
| 左旋肉碱 | 用于治疗功能障碍疾病 | 大肠杆菌 | 61.3 g/L | [ | |
| β-丙氨酸 | 生物体中泛酸生产的主要前体 | 大肠杆菌 | 43.12 g/L | [ | |
| 5-氨基戊酸酯 | 生产尼龙的结构单元 | 谷氨酸棒状杆菌 | 28 g/L | [ | |
| 5-氨基乙酰丙酸 | 用于多种癌症的光动力学诊断和治疗中 | 谷氨酸棒状杆菌 | 2.06 g/L | [ | |
| 转录调节与反馈抑制调节 | L-酪氨酸 | 食品和饲料添加剂 | 酿酒酵母 | 36.2 mg/L | [ |
| 2-氨基丁酸 | 抗结核性乙胺丁醇的重要前体 | 大肠杆菌 | 5.39 g/L | [ | |
| L-精氨酸 | 在食品和补充保健食品,制药和化妆品行业中有重要应用 | 谷氨酸棒状杆菌 | 61.9 g/L | [ | |
| L-亮氨酸 | 在刺激肌肉蛋白质合成和葡萄糖稳态中具有重要作用 | 谷氨酸棒状杆菌 | 5.7 g/L | [ | |
| L-苏氨酸 | 广泛用于食品、制药和化妆品行业 | 大肠杆菌 | 26.0 g/L | [ | |
| 转运系统的调节 | L-异亮氨酸 | 形成血红蛋白必需氨基酸,在调节血糖与能量方面起到关键作用 | 谷氨酸棒状杆菌 | 26.8 g/L | [ |
| 高丝氨酸 | 诱导免疫系统,增强植物对疾病的抵抗力 | 大肠杆菌W3110 | 39.54 g/L | [ | |
| L-半胱氨酸 | 在提高动物的免疫调节和促进动物生长发育方面起着重要作用 | 大肠杆菌 | 634.4 mg/L | [ | |
| L-蛋氨酸 | 用于脂肪肝,以及酒精等引起的肝损害 | 大肠杆菌 | 593.5 mg/L | [ | |
| L-瓜氨酸 | 对于保持肌肉和肝脏健康有重要作用 | 谷氨酸棒状杆菌 | 21 g/L | [ | |
| L-苏氨酸 | 促进人体发育和抗脂肪肝的药用效能 | 谷氨酸棒状杆菌 | 8.1 g/L | [ |
| 1 | Choi S, Song C W, Shin J H, et al. Biorefineries for the production of top building block chemicals and their derivatives[J]. Metabolic Engineering, 2015, 28: 223-239. |
| 2 | Piao X, Wang L, Lin B, et al. Metabolic engineering of Escherichia coli for production of L-aspartate and its derivative β-alanine with high stoichiometric yield[J]. Metabolic Engineering, 2019, 54: 244-254. |
| 3 | Mu W, Zhang T, Jiang B. An overview of biological production of L-theanine[J]. Biotechnology Advances, 2015, 33(3): 335-342. |
| 4 | Wang H, Liu W, Shi F, et al. Metabolic pathway engineering for high-level production of 5-hydroxytryptophan in Escherichia coli[J]. Metabolic Engineering, 2018, 48: 279-287. |
| 5 | Song Y, Li J, Shin H D, et al. Biotechnological production of alpha-keto acids: current status and perspectives[J]. Bioresource Technology, 2016, 219: 716-724. |
| 6 | Wendisch V F.Metabolic engineering advances and prospects for amino acid production[J].Metabolic Engineering, 2020, 58: 17-34. |
| 7 | D􀆳este M, Alvarado-Morales M, Angelidaki I. Amino acids production focusing on fermentation technologies — a review[J]. Biotechnology Advances,2018, 36(1): 14-25. |
| 8 | Farmer W R, Liao J C. Progress in metabolic engineering[J]. Current Opinion in Biotechnology, 1996, 7(2): 198-204. |
| 9 | Kusumoto I. Industrial production of L-glutamine[J]. The Journal of Nutrition, 2001, 131(9): 2552S-2555S. |
| 10 | Park J H, Lee S Y. Towards systems metabolic engineering of microorganisms for amino acid production[J]. Current Opinion in Biotechnology, 2008, 19(5): 454-460. |
| 11 | Chae T U, Choi S Y, Kim J W, et al. Recent advances in systems metabolic engineering tools and strategies[J]. Current Opinion in Biotechnology, 2017, 47: 67-82. |
| 12 | Kohlstedt M, Becker J, Wittmann C. Metabolic fluxes and beyond-systems biology understanding and engineering of microbial metabolism[J]. Applied Microbiology and Biotechnology, 2010, 88(5): 1065-1075. |
| 13 | Lee J W, Na D, Park J M, et al. Systems metabolic engineering of microorganisms for natural and non-natural chemicals[J]. Nature Chemical Biology, 2012, 8(6): 536-546. |
| 14 | Chen Z, Huang J, Wu Y, et al. Metabolic engineering of Corynebacterium glutamicum for the production of 3-hydroxypropionic acid from glucose and xylose[J]. Metabolic Engineering, 2017, 39: 151-158. |
| 15 | Kogure T, Kubota T, Suda M, et al. Metabolic engineering of Corynebacterium glutamicum for shikimate overproduction by growth-arrested cell reaction[J]. Metabolic Engineering, 2016, 38: 204-216. |
| 16 | Yao R, Shimizu K. Recent progress in metabolic engineering for the production of biofuels and biochemicals from renewable sources with particular emphasis on catabolite regulation and its modulation[J]. Process Biochemistry, 2013, 48(9): 1409-1417. |
| 17 | Xu J Z, Yu H B, Han M, et al. Metabolic engineering of glucose uptake systems in Corynebacterium glutamicum for improving the efficiency of L-lysine production[J]. Journal of Industrial Microbiology & Biotechnology, 2019, 46(7): 937-949. |
| 18 | Lindner S N, Knebel S, Pallerla S R, et al. Cg2091 encodes a polyphosphate/ATP-dependent glucokinase of Corynebacterium glutamicum[J]. Applied Microbiology and Biotechnology, 2010, 87(2): 703-713. |
| 19 | Lindner S N, Seibold G M, Henrich A, et al. Phosphotransferase system-independent glucose utilization in Corynebacterium glutamicum by inositol permeases and glucokinases[J]. Applied and Environmental Microbiology, 2011, 77(11): 3571-3581. |
| 20 | Zhang B, Gao G, Chu X H, et al. Metabolic engineering of Corynebacterium glutamicum S9114 to enhance the production of L-ornithine driven by glucose and xylose[J]. Bioresource Technology, 2019, 284: 204-213. |
| 21 | Klaffl S, Brocker M, Kalinowski J, et al. Complex regulation of the phosphoenolpyruvate carboxykinase gene pck and characterization of its GntR-type regulator IolR as a repressor of myo-inositol utilization genes in Corynebacterium glutamicum[J]. Journal of Bacteriology, 2013, 195(18): 4283-4296. |
| 22 | Vogt M, Haas S, Klaffl S, et al. Pushing product formation to its limit: metabolic engineering of Corynebacterium glutamicum for L-leucine overproduction[J]. Metabolic Engineering, 2014, 22: 40-52. |
| 23 | Fordjour E, Adipah F K, Zhou S, et al. Metabolic engineering of Escherichia coli BL21 (DE3) for de novo production of L-DOPA from D-glucose[J]. Microbial Cell Factories, 2019, 18(1): 74. |
| 24 | Patnaik R, Liao J C. Engineering of Escherichia coli central metabolism for aromatic metabolite production with near theoretical yield[J]. Applied and Environmental Microbiology, 1994, 60(11): 3903. |
| 25 | Báez J L, Bolívar F, Gosset G. Determination of 3-deoxy-D-arabino-heptulosonate 7-phosphate productivity and yield from glucose in Escherichia coli devoid of the glucose phosphotransferase transport system[J]. Biotechnology and Bioengineering, 2001, 73(6): 530-535. |
| 26 | Muñoz A J, Hernández-Chávez G, De Anda R, et al. Metabolic engineering of Escherichia coli for improving L-3,4-dihydroxyphenylalanine (L-DOPA) synthesis from glucose[J]. Journal of Industrial Microbiology & Biotechnology, 2011, 38(11): 1845. |
| 27 | Zou X, Guo L, Huang L, et al. Pathway construction and metabolic engineering for fermentative production of β-alanine in Escherichia coli[J]. Applied Microbiology and Biotechnology, 2020, 104(6): 2545-2559. |
| 28 | Glick B R. Metabolic load and heterologous gene expression[J]. Biotechnology Advances, 1995, 13(2): 247-261. |
| 29 | Koma D, Kishida T, Yamanaka H, et al. Escherichia coli chromosome-based T7-dependent constitutive overexpression system and its application to generating a phenylalanine producing strain[J]. Journal of Bioscience and Bioengineering, 2018, 126(5): 586-595. |
| 30 | Li Z J, Hong P H, Da Y Y, et al. Metabolic engineering of Escherichia coli for the production of L-malate from xylose[J]. Metabolic Engineering, 2018, 48: 25-32. |
| 31 | Ruan L, Li L, Zou D, et al. Metabolic engineering of Bacillus amyloliquefaciens for enhanced production of S-adenosylmethionine by coupling of an engineered S-adenosylmethionine pathway and the tricarboxylic acid cycle[J]. Biotechnology for Biofuels, 2019, 12(1): 211. |
| 32 | Smirnov S V, Kodera T, Samsonova N N, et al. Metabolic engineering of Escherichia coli to produce (2S, 3R, 4S)-4-hydroxyisoleucine[J]. Applied Microbiology and Biotechnology, 2010, 88(3): 719-726. |
| 33 | Frost J W. Altered glucose transport and shikimate pathway product yields in E.coli[J]. Biotechnology Progress, 2003, 19(5): 1450-1459. |
| 34 | Berry A. Improving production of aromatic compounds in Escherichia coli by metabolic engineering[J]. Trends in Biotechnology, 1996, 14(7): 250-256. |
| 35 | Marienhagen J. Metabolic engineering of Corynebacterium glutamicum for the metabolization of methanol[J]. Appl. Environ. Microbiol., 2015, 81(6): 2215-2225. |
| 36 | Liu H, Marsafari M, Wang F, et al. Engineering acetyl-CoA metabolic shortcut for eco-friendly production of polyketides triacetic acid lactone in Yarrowia lipolytica[J]. Metabolic Engineering, 2019, 56: 60-68. |
| 37 | Jansson C. Metabolic engineering of Cyanobacteria for direct conversion of CO2 to hydrocarbon biofuels[M]. Lüttge U, Beyschlag W, Büdel B, et al. Progress in Botany 73. Berlin, Heidelberg: Springer, 2012: 81-93. |
| 38 | Nguyen D T N, Lee O K, Lim C, et al. Metabolic engineering of type Ⅱ methanotroph, Methylosinus trichosporium OB3b, for production of 3-hydroxypropionic acid from methane via a malonyl-CoA reductase-dependent pathway[J]. Metabolic Engineering, 2020, 59: 142-150. |
| 39 | Gleizer S, Ben Nissan R, Bar On Y M, et al. Conversion of Escherichia coli to generate all biomass carbon from CO2[J]. Cell, 2019, 179(6): 1255-1263. |
| 40 | Wu G, Yan Q, Jones J A, et al. Metabolic burden: cornerstones in synthetic biology and metabolic engineering applications[J]. Trends in Biotechnology, 2016, 34(8): 652-664. |
| 41 | Li H, Wang B, Zhu L, et al. Metabolic engineering of Escherichia coli W3110 for L-homoserine production[J]. Process Biochemistry, 2016, 51(12): 1973-1983. |
| 42 | Zhang K, Li H, Cho K M, et al. Expanding metabolism for total biosynthesis of the nonnatural amino acid L-homoalanine[J]. Proceedings of the National Academy of Sciences, 2010, 107(14): 6234. |
| 43 | Ginesy M, Belotserkovsky J, Enman J, et al. Metabolic engineering of Escherichia coli for enhanced arginine biosynthesis[J]. Microbial Cell Factories, 2015, 14(1): 29. |
| 44 | Man Z, Xu M, Rao Z, et al. Systems pathway engineering of Corynebacterium crenatum for improved L-arginine production[J]. Scientific Reports, 2016, 6(1): 28629. |
| 45 | Zhan M, Kan B, Dong J, et al. Metabolic engineering of Corynebacterium glutamicum for improved L-arginine synthesis by enhancing NADPH supply[J]. Journal of Industrial Microbiology & Biotechnology, 2019, 46(1): 45-54. |
| 46 | Bommareddy R R, Chen Z, Rappert S, et al. A de novo NADPH generation pathway for improving lysine production of Corynebacterium glutamicum by rational design of the coenzyme specificity of glyceraldehyde 3-phosphate dehydrogenase[J]. Metabolic Engineering, 2014, 25: 30-37. |
| 47 | Lin Y, Sun X, Yuan Q, et al. Engineering bacterial phenylalanine 4-hydroxylase for microbial synthesis of human neurotransmitter precursor 5-hydroxytryptophan[J]. ACS Synthetic Biology, 2014, 3(7): 497-505. |
| 48 | Pribat A, Blaby I K, Lara Núñez A, et al. FolX and FolM are essential for tetrahydromonapterin synthesis in Escherichia coli and Pseudomonas aeruginosa[J]. J. Bacteriol., 2010, 192(2): 475-482. |
| 49 | Gao W, Sun H X, Xiao H, et al. Combining metabolomics and transcriptomics to characterize tanshinone biosynthesis in Salvia miltiorrhiza[J]. BMC Genomics, 2014, 15(1): 73. |
| 50 | Thiele I, Palsson B. A protocol for generating a high-quality genome-scale metabolic reconstruction[J]. Nature Protocols, 2010, 5(1): 93-121. |
| 51 | Amador Noguez D, Feng X J, Fan J, et al. Systems-level metabolic flux profiling elucidates a complete, bifurcated tricarboxylic acid cycle in Clostridium acetobutylicum[J]. Journal of Bacteriology, 2011, 193(23): 6805. |
| 52 | Venkataramanan K P, Min L, Hou S, et al. Complex and extensive post-transcriptional regulation revealed by integrative proteomic and transcriptomic analysis of metabolite stress response in Clostridium acetobutylicum[J]. Biotechnology for Biofuels, 2015, 8(1): 81. |
| 53 | Xu J Y, Xu Y, Chu X, et al. Protein acylation affects the artificial biosynthetic pathway for pinosylvin production in engineered E. coli[J]. ACS Chemical Biology, 2018, 13(5): 1200-1208. |
| 54 | Yoo M, Nguyen N P T, Soucaille P. Trends in systems biology for the analysis and engineering of Clostridium acetobutylicum metabolism[J]. Trends in Microbiology, 2020, 28(2): 118-140. |
| 55 | Cheah Y E, Xu Y, Sacco S A, et al. Systematic identification and elimination of flux bottlenecks in the aldehyde production pathway of Synechococcus elongatus PCC 7942[J]. Metabolic Engineering, 2020, 60: 56-65. |
| 56 | Maiti M K. Functional characterization of two structurally novel diacylglycerol acyltransferase 2 isozymes responsible for the enhanced production of stearate-rich storage lipid in Candida tropicalis SY005[J]. Plos One, 2014, 9(4): e94472. |
| 57 | Yuzawa S, Mirsiaghi M, Jocic R, et al. Short-chain ketone production by engineered polyketide synthases in Streptomyces albus[J]. Nature Communications, 2018, 9(1): 4569. |
| 58 | Bali A P, Lennox Hvenekilde D, Myling Petersen N, et al. Improved biotin, thiamine, and lipoic acid biosynthesis by engineering the global regulator IscR[J]. Metabolic Engineering, 2020, 60: 97-109. |
| 59 | Mundhada H, Schneider K, Christensen H B, et al. Engineering of high yield production of L-serine in Escherichia coli[J]. Biotechnology and Bioengineering, 2016, 113(4): 807-816. |
| 60 | Zhang B, Yu M, Zhou Y, et al. Systematic pathway engineering of Corynebacterium glutamicum S9114 for L-ornithine production[J]. Microbial Cell Factories, 2017, 16(1): 158. |
| 61 | Arense P, Bernal V, Charlier D, et al. Metabolic engineering for high yielding L(-)-carnitine production in Escherichia coli[J]. Microbial Cell Factories, 2013, 12(1): 56. |
| 62 | Rohles C M, Gießelmann G, Kohlstedt M, et al. Systems metabolic engineering of Corynebacterium glutamicum for the production of the carbon-5 platform chemicals 5-aminovalerate and glutarate[J]. Microbial Cell Factories, 2016, 15(1): 154. |
| 63 | Feng L, Zhang Y, Fu J, et al. Metabolic engineering of Corynebacterium glutamicum for efficient production of 5-aminolevulinic acid[J]. Biotechnology and Bioengineering, 2016, 113(6): 1284-1293. |
| 64 | Miscevic D, Srirangan K, Kefale T, et al. Heterologous production of 3-hydroxyvalerate in engineered Escherichia coli[J]. Metabolic Engineering, 2020, 61: 141-151. |
| 65 | Park J H, Lee K H, Kim T Y, et al. Metabolic engineering of Escherichia coli for the production of L-valine based on transcriptome analysis and in silico gene knockout simulation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(19): 7797-7802. |
| 66 | Lee K H, Park J H, Kim T Y, et al. Systems metabolic engineering of Escherichia coli for L-threonine production[J]. Molecular Systems Biology, 2007, 3(1): 149. |
| 67 | Fisher M A, Boyarskiy S, Yamada M R, et al. Enhancing tolerance to short-chain alcohols by engineering the Escherichia coli AcrB efflux pump to secrete the non-native substrate n-butanol[J]. ACS Synthetic Biology, 2014, 3(1): 30-40. |
| 68 | Alves T C, Pongratz R L, Zhao X, et al. Integrated, step-wise, mass-isotopomeric flux analysis of the TCA cycle[J]. Cell Metabolism, 2015, 22(5): 936-947. |
| 69 | Quek L E, Krycer J R, Ohno S, et al. Dynamic 13C flux analysis captures the reorganization of adipocyte glucose metabolism in response to insulin[J]. iScience, 2020, 23(2): 100855. |
| 70 | Farmer W R, Liao J C. Precursor balancing for metabolic engineering of lycopene production in Escherichia coli[J]. Biotechnology Progress, 2001, 17(1): 57-61. |
| 71 | Künzler M, Paravicini G, Egli C M, et al. Cloning, primary structure and regulation of the ARO4 gene, encoding the tyrosine-inhibited 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase from Saccharomyces cerevisiae[J]. Gene, 1992, 113(1): 67-74. |
| 72 | Brown J F, Dawes I W. Regulation of chorismate mutase in Saccharomyces cerevisiae[J]. Molecular and General Genetics MGG, 1990, 220(2): 283-288. |
| 73 | Luttik M A H, Vuralhan Z, Suir E, et al. Alleviation of feedback inhibition in Saccharomyces cerevisiae aromatic amino acid biosynthesis: quantification of metabolic impact[J]. Metabolic Engineering, 2008, 10(3): 141-153. |
| 74 | Xu J M, Li J Q, Zhang B, et al. Fermentative production of the unnatural amino acid L-2-aminobutyric acid based on metabolic engineering[J]. Microbial Cell Factories, 2019, 18(1): 43. |
| 75 | Park S H, Kim H U, Kim T Y, et al. Metabolic engineering of Corynebacterium glutamicum for L-arginine production[J]. Nature Communications, 2014, 5(1): 4618. |
| 76 | Xu M, Rao Z, Dou W, et al. The role of ARGR repressor regulation on L-arginine production in Corynebacterium crenatum[J]. Applied Biochemistry and Biotechnology, 2013, 170(3): 587-597. |
| 77 | Ding Z, Fang Y, Zhu L, et al. Deletion of arcA, iclR, and tdcC in Escherichia coli to improve L-threonine production[J]. Biotechnology and Applied Biochemistry, 2019, 66(5): 794-807. |
| 78 | Huang J F, Liu Z Q, Jin L Q, et al. Metabolic engineering of Escherichia coli for microbial production of L-methionine[J]. Biotechnology and Bioengineering, 2017, 114(4): 843-851. |
| 79 | Grove A. Regulation of metabolic pathways by MarR family transcription factors[J]. Computational and Structural Biotechnology Journal, 2017, 15: 366-371. |
| 80 | Swint-Kruse L, Matthews K S. Allostery in the LacI/GalR family: variations on a theme[J]. Current Opinion in Microbiology, 2009, 12(2): 129-137. |
| 81 | Subrata B, Susobhan C, Dilip S, et al. Dynamical perspective of protein-DNA interaction[J]. Biomolecular Concepts, 2014, 5(1): 21-43. |
| 82 | Wray L V, Ferson A E, Fisher S H. Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors including CodY, GlnR, TnrA, and Spo0H[J]. Journal of Bacteriology, 1997, 179(17): 5494. |
| 83 | Loper J C. Multiple regulatory elements control expression of the gene encoding the Saccharomyces cerevisiae cytochrome P450, lanosterol 14 alpha-demethylase (ERG11)[J]. Journal of Biological Chemistry, 1992, 267(3): 2046. |
| 84 | Yin L, Shi F, Hu X, et al. Increasing L-isoleucine production in Corynebacterium glutamicum by overexpressing global regulator Lrp and two-component export system BrnFE[J]. Journal of Applied Microbiology, 2013, 114(5): 1369-1377. |
| 85 | Rendić M. Amino acids in animal nutrition[J]. Amino Acids in Animal Nutrition, 2003, 64(1): 488. |
| 86 | Park S, Imlay J A. High levels of intracellular cysteine promote oxidative DNA damage by driving the fenton reaction[J]. Journal of Bacteriology, 2003, 185(6): 1942. |
| 87 | Wei L, Wang H, Xu N, et al. Metabolic engineering of Corynebacterium glutamicum for L-cysteine production[J]. Applied Microbiology and Biotechnology, 2019, 103(3): 1325-1338. |
| 88 | Lubitz D, Jorge J M P, Pérez-García F, et al. Roles of export genes cgmA and lysE for the production of L-arginine and L-citrulline by Corynebacterium glutamicum[J]. Applied Microbiology and Biotechnology, 2016, 100(19): 8465-8474. |
| 89 | Simic P, Willuhn J, Sahm H, et al. Identification of glyA (encoding serine hydroxymethyltransferase) and its use together with the exporter ThrE to increase L-threonine accumulation by Corynebacterium glutamicum[J]. Appl. Environ. Microbiol., 2002, 68(7): 3321-3327. |
| 90 | Matano C, Uhde A, Youn J W, et al. Engineering of Corynebacterium glutamicum for growth and L-lysine and lycopene production from N-acetyl-glucosamine[J]. Applied Microbiology and Biotechnology, 2014, 98(12): 5633-5643. |
| 91 | Qin J, Zhou Y J, Krivoruchko A, et al. Modular pathway rewiring of Saccharomyces cerevisiae enables high-level production of L-ornithine[J]. Nature Communications, 2015, 6(1): 8224. |
| 92 | Smolke C D. Building outside of the box: iGEM and the biobricks foundation[J]. Nature Biotechnology, 2009, 27(12): 1099-1102. |
| 93 | Zhou L B, Zeng A P. Exploring lysine riboswitch for metabolic flux control and improvement of L-lysine synthesis in Corynebacterium glutamicum[J]. ACS Synthetic Biology, 2015, 4(6): 729-734. |
| 94 | Lee J H, Wendisch V F. Production of amino acids — genetic and metabolic engineering approaches[J]. Bioresource Technology, 2017, 245: 1575-1587. |
| 95 | Becker J, Wittmann C. Systems and synthetic metabolic engineering for amino acid production — the heartbeat of industrial strain development[J]. Current Opinion in Biotechnology, 2012, 23(5): 718-726. |
| [1] | 刘昕, 戈钧, 李春. 光驱动微生物杂合系统提高生物制造水平[J]. 化工学报, 2023, 74(1): 330-341. |
| [2] | 毕浩然, 张洋, 王凯, 徐晨晨, 霍奕影, 陈必强, 谭天伟. 微生物制造绿色化学品研究进展[J]. 化工学报, 2023, 74(1): 1-13. |
| [3] | 刘雪, 张莉娟, 赵广荣. 大肠杆菌偏利共培养系统合成大豆苷元[J]. 化工学报, 2022, 73(9): 4015-4024. |
| [4] | 孙怡, 张腾, 吕波, 李春. 胞内生物传感器提高微生物细胞工厂的精细调控[J]. 化工学报, 2022, 73(2): 521-534. |
| [5] | 王靖楠, 庞建, 秦磊, 郭超, 吕波, 李春, 王超. 丁烯基多杀菌素高产菌株的选育和改造策略[J]. 化工学报, 2022, 73(2): 566-576. |
| [6] | 王欣慧, 王颖, 姚明东, 肖文海. 维生素A生物合成的研究进展[J]. 化工学报, 2022, 73(10): 4311-4323. |
| [7] | 周武林, 高惠芳, 吴玉玲, 张显, 徐美娟, 杨套伟, 邵明龙, 饶志明. 重组酿酒酵母生物合成菜油甾醇[J]. 化工学报, 2021, 72(8): 4314-4324. |
| [8] | 高子熹, 郭树奇, 费强. 生物转化温室气体生产单细胞蛋白的研究进展[J]. 化工学报, 2021, 72(6): 3202-3214. |
| [9] | 王欣, 赵鹏, 李清扬, 田平芳. 半导体合成生物学的研究进展[J]. 化工学报, 2021, 72(5): 2426-2435. |
| [10] | 毛金竹, 肖淑玲, 杨智淳, 王孝宇, 张诗, 陈俊宏, 谢佶晟, 陈福德, 黄子诺, 冯天宇, 张瑷珲, 方柏山. 合成生物学在农残检测领域的应用[J]. 化工学报, 2021, 72(5): 2413-2425. |
| [11] | 王炼, 吴迪, 周景文. 木脂素的生物合成及其微生物法生产的研究进展[J]. 化工学报, 2021, 72(1): 320-333. |
| [12] | 赵贞尧, 张保财, 李锋, 宋浩. 产电细胞的合成生物学设计构建[J]. 化工学报, 2021, 72(1): 468-482. |
| [13] | 王凯峰, 王金鹏, 韦萍, 纪晓俊. 代谢工程改造解脂耶氏酵母生产脂肪酸及其衍生物[J]. 化工学报, 2021, 72(1): 351-365. |
| [14] | 徐静, 由紫暄, 张君奇, 陈正, 吴德光, 李锋, 宋浩. 合成生物学方法改造电活性生物膜研究进展[J]. 化工学报, 2020, 71(9): 3950-3962. |
| [15] | 秦磊, 俞杰, 宁小钰, 孙文涛, 李春. 合成生物系统构建与绿色生物“智”造[J]. 化工学报, 2020, 71(9): 3979-3994. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备 11010102001995号
京公网安备 11010102001995号