化工学报 ›› 2021, Vol. 72 ›› Issue (8): 4314-4324.DOI: 10.11949/0438-1157.20201686
周武林( ),高惠芳,吴玉玲,张显,徐美娟,杨套伟,邵明龙(
),高惠芳,吴玉玲,张显,徐美娟,杨套伟,邵明龙( ),饶志明(
),饶志明( )
)
收稿日期:2020-11-25
修回日期:2021-01-19
出版日期:2021-08-05
发布日期:2021-08-05
通讯作者:
邵明龙,饶志明
作者简介:周武林(1997—),男,硕士研究生, 基金资助:
Wulin ZHOU( ),Huifang GAO,Yuling WU,Xian ZHANG,Meijuan XU,Taowei YANG,Minglong SHAO(
),Huifang GAO,Yuling WU,Xian ZHANG,Meijuan XU,Taowei YANG,Minglong SHAO( ),Zhiming RAO(
),Zhiming RAO( )
)
Received:2020-11-25
Revised:2021-01-19
Online:2021-08-05
Published:2021-08-05
Contact:
Minglong SHAO,Zhiming RAO
摘要:
菜油甾醇作为甾体药物(孕酮、雄烯二酮、氢化可的松等)的重要合成前体已受到国内外研究学者的广泛关注。首先通过生物信息学分析,筛选了10种不同来源的7-脱氢胆固醇还原酶DHCR7,并采用CRISPR/Cas9基因编辑技术将酿酒酵母(Saccharomyces cerevisiae)内源的ERG5基因替换成不同来源的DHCR7基因,构建了菜油甾醇合成菌株。结果发现整合来源于Pangasianodon hypophthalmus DHCR7的菌株Zw507表现出最高的菜油甾醇的产量216.93 mg/L。进一步筛选了10种酵母内源启动强度较强的启动子来与PhDHCR7基因进行组合,结果显示以TEF1p为启动子时菜油甾醇的产量最高可达253.35 mg/L。为了进一步提高菜油甾醇产量,增加了DHCR7表达盒在酵母基因组上的拷贝数。当拷贝数为3个时,菜油甾醇的产量达到最高302.27 mg/L。最终,通过5 L发酵罐进行补料分批发酵,实现了916.88 mg/L菜油甾醇产量。该菌株可作为后续甾体药物生物合成的优良底盘细胞。
中图分类号:
周武林, 高惠芳, 吴玉玲, 张显, 徐美娟, 杨套伟, 邵明龙, 饶志明. 重组酿酒酵母生物合成菜油甾醇[J]. 化工学报, 2021, 72(8): 4314-4324.
Wulin ZHOU, Huifang GAO, Yuling WU, Xian ZHANG, Meijuan XU, Taowei YANG, Minglong SHAO, Zhiming RAO. Engineering of Saccharomyces cerevisiae for biosynthesis of campesterol[J]. CIESC Journal, 2021, 72(8): 4314-4324.
| 菌株和质粒 | 性质 | 来源 |
|---|---|---|
菌株 S.cerevisiae | ||
| GTy23 | ERG9::KanMX_PCTR3-ERG9LEU2-3, 112::His3MX6_PGAL1 ERG19/PGAL10-ERG8 URA3-52::URA3_PGAL1-mvaS (A110G)/PGAL10-mvaE(CO) HIS3Δ1::hphMX4_PGAL1ERG12/PGAL10-IDI1 | Keasling教授惠赠[ |
| Zw501 | GTy23 (ura3-52 prototrophy removed for use of Cas9 system) | 本研究构建 |
| Zw502 | Zw501 (敲除了erg5) | 本研究构建 |
| Zw503 | Zw501 (erg5::GAL1p-DrDHCR7-ADH1t) | 本研究构建 |
| Zw504 | Zw501 (erg5::GAL1p-CaDHCR7-ADH1t) | 本研究构建 |
| Zw505 | Zw501 (erg5::GAL1p-TtDHCR7-ADH1t) | 本研究构建 |
| Zw506 | Zw501 (erg5::GAL1p-CmDHCR7-ADH1t) | 本研究构建 |
| Zw507 | Zw501 (erg5::GAL1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw508 | Zw501 (erg5::GAL1p-CcDHCR7-ADH1t) | 本研究构建 |
| Zw509 | Zw501 (erg5::GAL1p-LrDHCR7-ADH1t) | 本研究构建 |
| Zw510 | Zw501 (erg5::GAL1p-CgDHCR7-ADH1t) | 本研究构建 |
| Zw511 | Zw501 (erg5::GAL1p-XmDHCR7-ADH1t) | 本研究构建 |
| Zw512 | Zw501 (erg5::GAL1p-AmDHCR7-ADH1t) | 本研究构建 |
| Zw513 | Zw501 (erg5::TPI1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw514 | Zw501 (erg5::TEF1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw515 | Zw501 (erg5::TDH3p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw516 | Zw501 (erg5::TEF2p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw517 | Zw501 (erg5::PGK1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw518 | Zw501 (erg5::GPM1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw519 | Zw501 (erg5::GPD1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw520 | Zw501 (erg5::TDH2p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw521 | Zw501 (erg5::ACT1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw522 | Zw514 (1114a::TEF1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw523 | Zw522 (607b::TEF1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw524 | Zw523 (911b::TEF1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw525 | Zw524 (1014a::TEF1p-PhDHCR7-ADH1t) | 本研究构建 |
| Escherichia coli | ||
| DH5α | 用于本研究中质粒的构建与克隆 |
表1 本研究所有菌株和质粒
Table 1 Strains and plasmids used in this study
| 菌株和质粒 | 性质 | 来源 |
|---|---|---|
菌株 S.cerevisiae | ||
| GTy23 | ERG9::KanMX_PCTR3-ERG9LEU2-3, 112::His3MX6_PGAL1 ERG19/PGAL10-ERG8 URA3-52::URA3_PGAL1-mvaS (A110G)/PGAL10-mvaE(CO) HIS3Δ1::hphMX4_PGAL1ERG12/PGAL10-IDI1 | Keasling教授惠赠[ |
| Zw501 | GTy23 (ura3-52 prototrophy removed for use of Cas9 system) | 本研究构建 |
| Zw502 | Zw501 (敲除了erg5) | 本研究构建 |
| Zw503 | Zw501 (erg5::GAL1p-DrDHCR7-ADH1t) | 本研究构建 |
| Zw504 | Zw501 (erg5::GAL1p-CaDHCR7-ADH1t) | 本研究构建 |
| Zw505 | Zw501 (erg5::GAL1p-TtDHCR7-ADH1t) | 本研究构建 |
| Zw506 | Zw501 (erg5::GAL1p-CmDHCR7-ADH1t) | 本研究构建 |
| Zw507 | Zw501 (erg5::GAL1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw508 | Zw501 (erg5::GAL1p-CcDHCR7-ADH1t) | 本研究构建 |
| Zw509 | Zw501 (erg5::GAL1p-LrDHCR7-ADH1t) | 本研究构建 |
| Zw510 | Zw501 (erg5::GAL1p-CgDHCR7-ADH1t) | 本研究构建 |
| Zw511 | Zw501 (erg5::GAL1p-XmDHCR7-ADH1t) | 本研究构建 |
| Zw512 | Zw501 (erg5::GAL1p-AmDHCR7-ADH1t) | 本研究构建 |
| Zw513 | Zw501 (erg5::TPI1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw514 | Zw501 (erg5::TEF1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw515 | Zw501 (erg5::TDH3p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw516 | Zw501 (erg5::TEF2p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw517 | Zw501 (erg5::PGK1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw518 | Zw501 (erg5::GPM1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw519 | Zw501 (erg5::GPD1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw520 | Zw501 (erg5::TDH2p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw521 | Zw501 (erg5::ACT1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw522 | Zw514 (1114a::TEF1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw523 | Zw522 (607b::TEF1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw524 | Zw523 (911b::TEF1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw525 | Zw524 (1014a::TEF1p-PhDHCR7-ADH1t) | 本研究构建 |
| Escherichia coli | ||
| DH5α | 用于本研究中质粒的构建与克隆 |
| 引物 | 序列(5′→3′) |
|---|---|
| F-up-erg5 (delet) | TGGGAATACTGTACCAGATAATCAAACAT |
| R-up-erg5 (delet) | CAAAGTTCTGTTTTTCCCCATTTGTTAAAAGGTATTTATTGTCTATTGGAATAGC |
| F-down-erg5 (delet) | ATAAATACCTTTTAACAAATGGGGAAAAACAGAACTTTGTCCAGAC |
| R-down-erg5 (delet) | TGACAGTGACGAACGCTTCAG |
| F-erg5 | ATGAGTTCTGTCGCAGAAAATATAATAC |
| R-erg5 | TTATTCGAAGACTTCTCCAGTAATTGGG |
| F-up-erg5 | TGGGAATACTGTACCAGATAATCAAACATTAAA |
| R-up-erg5 | TGTTTATACGCTATTATCAGCCAATTTGTTAAAAGGTATTTATTGTCTATTGGAATAGCA |
| F-GAL1p | CCAATAGACAATAAATACCTTTTAACAAATTGGCTGATAATAGCGTATAAACAATGCA |
| R- GAL1p | TAACTCTATCAGAAGCCATCATTTTGTAATTAAAACTTAGATTAGATTGCTATGCTTTCT |
| F-DrDHCR7 | ATCTAAGTTTTAATTACAAAATGATGGCTTCTGATAGAGTTAGAAAAAG |
| R-DrDHCR7 | ATAAATCATAAGAAATTCGCTTAAAAAATATTTGGCAACAATCTATAAGAAACAGCAG |
| F-PhDHCR7 | ATCTAAGTTTTAATTACAAAATGTCTACCTCTGAAGGTGTTAGAAAAAG |
| R-PhDHCR7 | ATAAATCATAAGAAATTCGCTTAAAAAATACCTGGCAACAATCTATATCTAACGG |
| F-LrDHCR7 | ATCTAAGTTTTAATTACAAAATGGGTAGAGTTAAATGGAGATCTATTACC |
| R-LrDHCR7 | ATAAATCATAAGAAATTCGCTTAAAAAATATTTGGCAACAATCTATATGGAACAGC |
| F-CaDHCR7 | ATCTAAGTTTTAATTACAAAATGACCACCGCTGATGCT |
| R-CaDHCR7 | ATAAATCATAAGAAATTCGCTTAAAAAATATTTGGCAACAATCTATATGGAACAGC |
| F-XmDHCR7 | ATCTAAGTTTTAATTACAAAATGGAATCTACTAGAAGAAGACCTCAAC |
| R-XmDHCR7 | TTTTAAAACCTAAGAGTCACTGAACAGACCAGGAATCAATCTATGTG |
| F-CmDHCR7 | ATCTAAGTTTTAATTACAAAATGTCTAATCCATTTTTTGAATTACATCACCAG |
| R-CmDHCR7 | TTAATAATAAAAATCATAAAAAATAATCCAGGTAAAAGCCTCTGTGG |
| F-CgDHCR7 | ATCTAAGTTTTAATTACAAAATGATCGTATCAACTTGGTCCGG |
| R-CgDHCR7 | ACACTTATTTTTTTTATAACAAAGACACCAGGAATTAATCTGTCTGG |
| F-AmDHCR7 | ATCTAAGTTTTAATTACAAAATGTCTACAACCGAGAGTGTAAGG |
| R-AmDHCR7 | ACACTTATTTTTTTTATAACAAAAATCCCAGGTAACAATCTCTGC |
| F-TtDHCR7 | ATCTAAGTTTTAATTACAAAATGGCTTGTGATCAATATCAATGTTCT |
| R-TtDHCR7 | ATAAATCATAAGAAATTCGCTTAAAAAATACCTGGCAACAATCTATATGGAACAG |
| F-CcDHCR7 | ATCTAAGTTTTAATTACAAAATGACTACTGCAGATGCCGTC |
| R-CcDHCR7 | ATAAATCATAAGAAATTCGCGAAGATGTTAGGTAACAGTCTATAAGGCAC |
| F-ADH1t | TTTTTAAGCGAATTTCTTATGATTTATGATTTTTATTATTAAATAAGTTATAAAAAAAA |
| R- ADH1t | TGTCTGGACAAAGTTCTGTTTTTCCCCAGGAGTTAGCATATCTACAATTGGGTGAA |
| F-down-erg5 | CCCAATTGTAGATATGCTAACTCCTGGGGAAAAACAGAACTTTGTCCA |
| R-down-erg5 | TGACAGTGACGAACGCTTCAG |
| F-ACT1-qPCR | CTCCGTCTGGATTGGTGGTT |
| R-ACT1-qPCR | ACTTGTGGTGAACGATAGATGG |
| F-PhDHCR7-qPCR | TGTGATAGGCAGAGGCAAGAAT |
| R-PhDHCR7-qPCR | GGTGGTATAACTGGCGACGAT |
表2 本研究所用引物
Table 2 Primers used in this study
| 引物 | 序列(5′→3′) |
|---|---|
| F-up-erg5 (delet) | TGGGAATACTGTACCAGATAATCAAACAT |
| R-up-erg5 (delet) | CAAAGTTCTGTTTTTCCCCATTTGTTAAAAGGTATTTATTGTCTATTGGAATAGC |
| F-down-erg5 (delet) | ATAAATACCTTTTAACAAATGGGGAAAAACAGAACTTTGTCCAGAC |
| R-down-erg5 (delet) | TGACAGTGACGAACGCTTCAG |
| F-erg5 | ATGAGTTCTGTCGCAGAAAATATAATAC |
| R-erg5 | TTATTCGAAGACTTCTCCAGTAATTGGG |
| F-up-erg5 | TGGGAATACTGTACCAGATAATCAAACATTAAA |
| R-up-erg5 | TGTTTATACGCTATTATCAGCCAATTTGTTAAAAGGTATTTATTGTCTATTGGAATAGCA |
| F-GAL1p | CCAATAGACAATAAATACCTTTTAACAAATTGGCTGATAATAGCGTATAAACAATGCA |
| R- GAL1p | TAACTCTATCAGAAGCCATCATTTTGTAATTAAAACTTAGATTAGATTGCTATGCTTTCT |
| F-DrDHCR7 | ATCTAAGTTTTAATTACAAAATGATGGCTTCTGATAGAGTTAGAAAAAG |
| R-DrDHCR7 | ATAAATCATAAGAAATTCGCTTAAAAAATATTTGGCAACAATCTATAAGAAACAGCAG |
| F-PhDHCR7 | ATCTAAGTTTTAATTACAAAATGTCTACCTCTGAAGGTGTTAGAAAAAG |
| R-PhDHCR7 | ATAAATCATAAGAAATTCGCTTAAAAAATACCTGGCAACAATCTATATCTAACGG |
| F-LrDHCR7 | ATCTAAGTTTTAATTACAAAATGGGTAGAGTTAAATGGAGATCTATTACC |
| R-LrDHCR7 | ATAAATCATAAGAAATTCGCTTAAAAAATATTTGGCAACAATCTATATGGAACAGC |
| F-CaDHCR7 | ATCTAAGTTTTAATTACAAAATGACCACCGCTGATGCT |
| R-CaDHCR7 | ATAAATCATAAGAAATTCGCTTAAAAAATATTTGGCAACAATCTATATGGAACAGC |
| F-XmDHCR7 | ATCTAAGTTTTAATTACAAAATGGAATCTACTAGAAGAAGACCTCAAC |
| R-XmDHCR7 | TTTTAAAACCTAAGAGTCACTGAACAGACCAGGAATCAATCTATGTG |
| F-CmDHCR7 | ATCTAAGTTTTAATTACAAAATGTCTAATCCATTTTTTGAATTACATCACCAG |
| R-CmDHCR7 | TTAATAATAAAAATCATAAAAAATAATCCAGGTAAAAGCCTCTGTGG |
| F-CgDHCR7 | ATCTAAGTTTTAATTACAAAATGATCGTATCAACTTGGTCCGG |
| R-CgDHCR7 | ACACTTATTTTTTTTATAACAAAGACACCAGGAATTAATCTGTCTGG |
| F-AmDHCR7 | ATCTAAGTTTTAATTACAAAATGTCTACAACCGAGAGTGTAAGG |
| R-AmDHCR7 | ACACTTATTTTTTTTATAACAAAAATCCCAGGTAACAATCTCTGC |
| F-TtDHCR7 | ATCTAAGTTTTAATTACAAAATGGCTTGTGATCAATATCAATGTTCT |
| R-TtDHCR7 | ATAAATCATAAGAAATTCGCTTAAAAAATACCTGGCAACAATCTATATGGAACAG |
| F-CcDHCR7 | ATCTAAGTTTTAATTACAAAATGACTACTGCAGATGCCGTC |
| R-CcDHCR7 | ATAAATCATAAGAAATTCGCGAAGATGTTAGGTAACAGTCTATAAGGCAC |
| F-ADH1t | TTTTTAAGCGAATTTCTTATGATTTATGATTTTTATTATTAAATAAGTTATAAAAAAAA |
| R- ADH1t | TGTCTGGACAAAGTTCTGTTTTTCCCCAGGAGTTAGCATATCTACAATTGGGTGAA |
| F-down-erg5 | CCCAATTGTAGATATGCTAACTCCTGGGGAAAAACAGAACTTTGTCCA |
| R-down-erg5 | TGACAGTGACGAACGCTTCAG |
| F-ACT1-qPCR | CTCCGTCTGGATTGGTGGTT |
| R-ACT1-qPCR | ACTTGTGGTGAACGATAGATGG |
| F-PhDHCR7-qPCR | TGTGATAGGCAGAGGCAAGAAT |
| R-PhDHCR7-qPCR | GGTGGTATAACTGGCGACGAT |
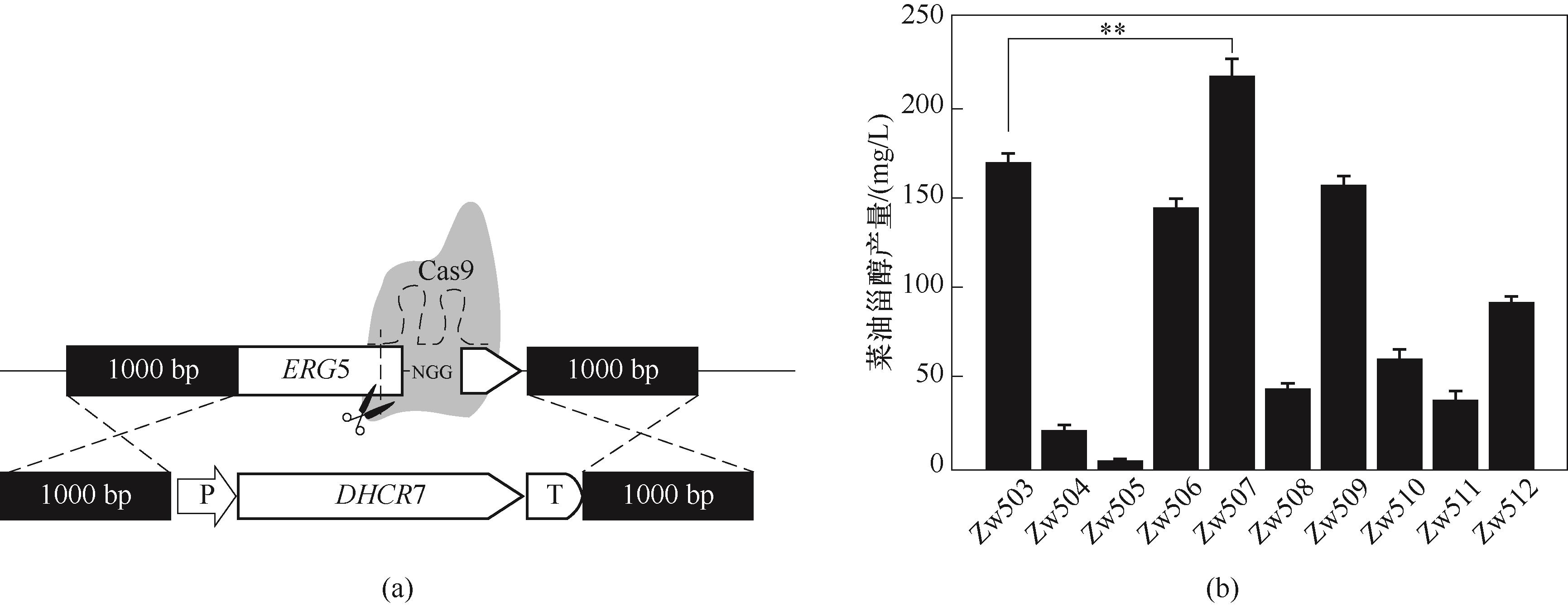
图3 重组菌株的构建(a)以及不同物种来源的DHCR7菜油甾醇产量(b)(**与对照菌株比较p≤0.01;由t检验确定,下同)
Fig.3 The construction of recombinant strains (a) and the production of DHCR7 campesterol from different species (b)( **p≤0.01 compared with the control strain; as determined by t test. The same below)
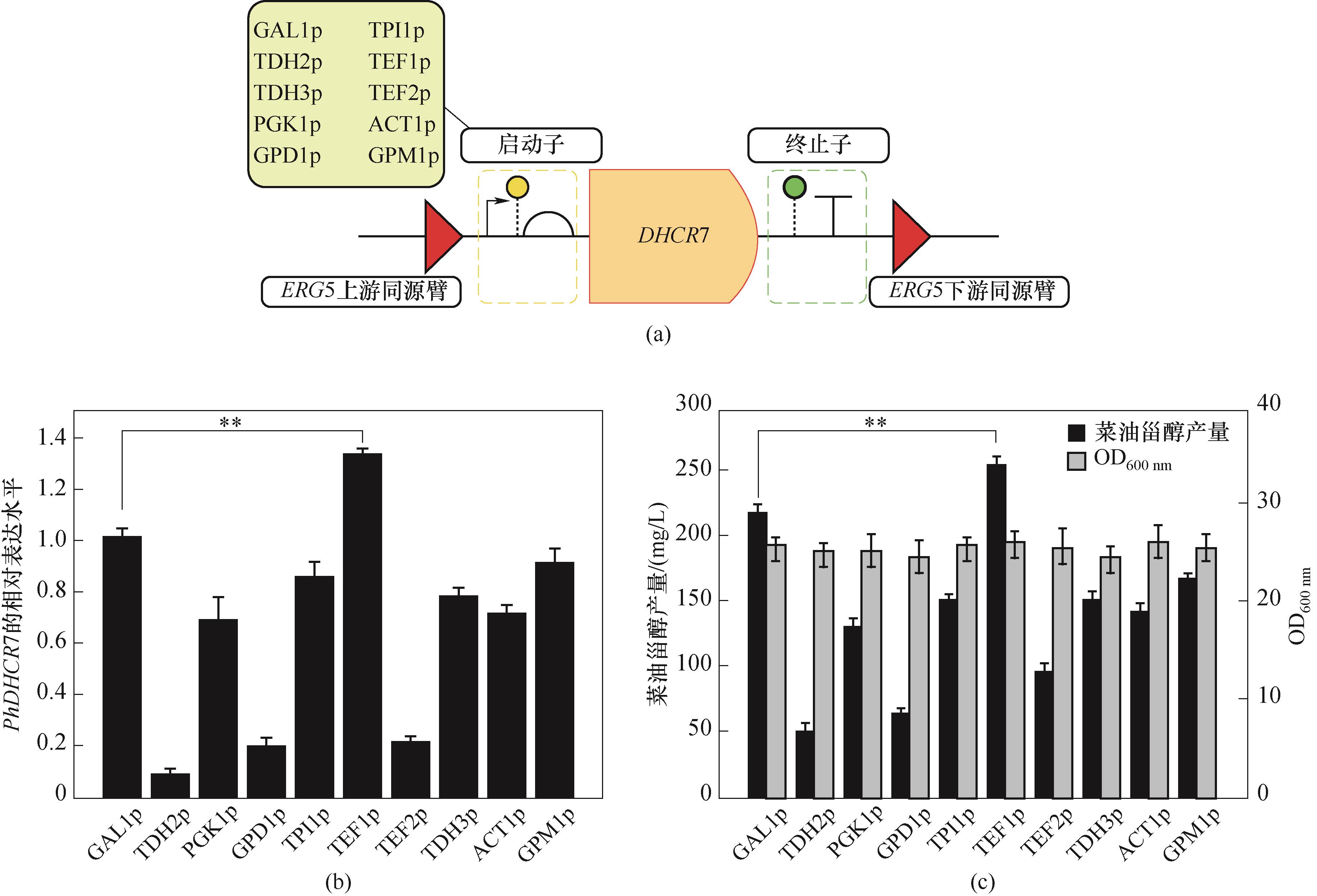
图4 不同启动子与PhDHCR7基因组合的示意图(a)、PhDHCR7表达量(b)以及菜油甾醇产量(c)
Fig.4 Schematic diagram (a), PhDHCR7 expression (b) and campesterol production (c) of combinations of different promoters and DHCR7 genes (**p<0.01)
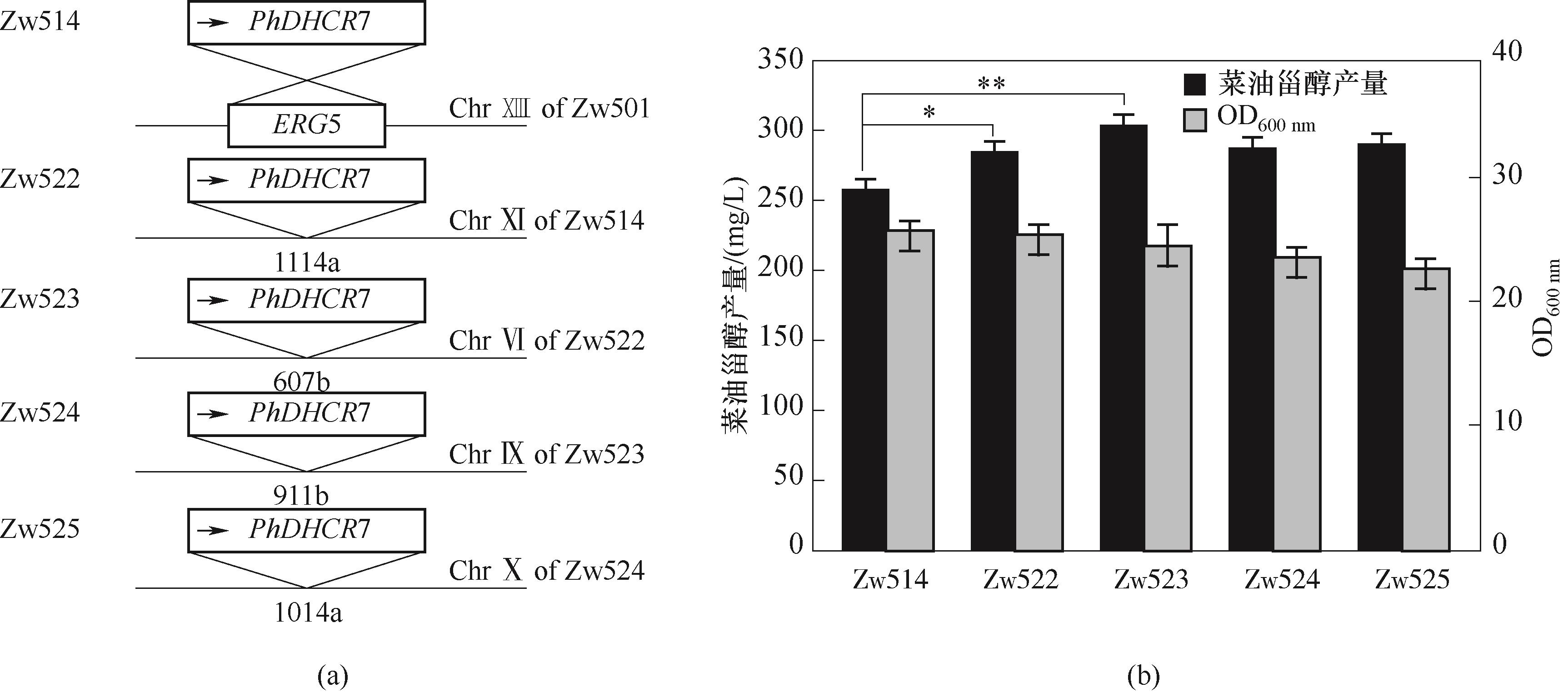
图5 PhDHCR7表达盒的多重拷贝数菌株的示意图(a)以及菜油甾醇产量(b)
Fig.5 Schematic diagram (a) and campesterol production (b) of the multiple copy number strain of PhDHCR7 expression cassette (**p<0.01, *p<0.05)
| 1 | Duport C, Spagnoli R, Degryse E, et al. Self-sufficient biosynthesis of pregnenolone and progesterone in engineered yeast[J]. Nature Biotechnology, 1998, 16 (2): 186-189. |
| 2 | Szczebara F M, Chandelier C, Villeret C, et al. Total biosynthesis of hydrocortisone from a simple carbon source in yeast[J]. Nature Biotechnology, 2003, 21 (2): 143-149. |
| 3 | Tsukagoshi Y, Suzuki H, Seki H, et al. Ajuga Δ24-sterol reductase catalyzes the direct reductive conversion of 24-methylenecholesterol to campesterol[J]. J. Biol. Chem., 2016, 291 (15): 8189-8198. |
| 4 | Zhao Y, Shen Y, Ma S, et al. Production of 5α-androstene-3,17-dione from phytosterols by co-expression of 5α-reductase and glucose-6-phosphate dehydrogenase in engineered Mycobacterium neoaurum[J]. Green Chemistry, 2019, 21 (7): 1809-1815. |
| 5 | Donova M V. Steroid bioconversions[J]. Methods Mol. Biol., 2017, 1645: 1-13. |
| 6 | Yao K, Xu L Q,Wang F Q, et al. Characterization and engineering of 3-ketosteroid-Δ1-dehydrogenase and 3-ketosteroid-9α-hydroxylase in Mycobacterium neoaurum ATCC 25795 to produce 9α-hydroxy-4-androstene-3,17-dione through the catabolism of sterols[J]. Metab. Eng., 2014, 24: 181-191. |
| 7 | Donova M V, Egorova O V. Microbial steroid transformations: current state and prospects[J]. Appl. Microbiol. Biotechnol., 2012, 94 (6): 1423-1447. |
| 8 | 张丽青.微生物转化在甾体药物合成中的应用[J].医药工业,1985, (1): 37-41. |
| Zhang L Q. Application of microbial transformation in the synthesis of steroid drugs [J]. Chinese Journal of Pharmaceuticals, 1985, (1): 37-41. | |
| 9 | Chen J, Fan F, Qu G, et al. Identification of Absidia orchidis steroid 11β-hydroxylation system and its application in engineering Saccharomyces cerevisiae for one-step biotransformation to produce hydrocortisone[J]. Metab. Eng., 2020, 57: 31-42. |
| 10 | Zhang W, Shao M, Rao Z, et al. Bioconversion of 4-androstene-3,17-dione to androst-1,4-diene-3,17-dione by recombinant Bacillus subtilis expressing ksdd gene encoding 3-ketosteroid-Δ1-dehydrogenase from Mycobacterium neoaurum JC-12[J]. The Journal of Steroid Biochemistry and Molecular Biology, 2013, 135: 36-42. |
| 11 | 吴玉玲,邵明龙,周武林, 等. 重组大肠杆菌表达17β-羟基类固醇脱氢酶全细胞催化合成宝丹酮的研究[J]. 化工学报, 2020, 71(7): 3229-3237. |
| Wu Y L, Shao M L, Zhou W L, et al. Study on catalytic synthesis of boldenone by recombinant Escherichia coli expressing 17β-hydroxysteroid dehydrogenase[J]. CIESC Journal, 2020, 71(7): 3229-3237. | |
| 12 | Ferreira R, Teixeira P G, Gossing M, et al. Metabolic engineering of Saccharomyces cerevisiae for overproduction of triacylglycerols[J]. Metab. Eng. Commun., 2018, 6: 22-27. |
| 13 | DiCarlo J E, Norville J E, Mali P, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems[J]. Nucleic Acids Res., 2013, 41 (7): 4336-4343. |
| 14 | Jakočiūnas T, Bonde I, Herrgård M, et al. Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae[J]. Metab. Eng., 2015, 28: 213-222. |
| 15 | Zalatan J G, Lee M E, Almeida R, et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds[J]. Cell, 2015, 160 (1): 339-350. |
| 16 | Lecain E, Chenivesse X, Spagnoli R, et al. Cloning by metabolic interference in yeast and enzymatic characterization of Arabidopsis thaliana sterol Δ7-reductase[J]. J. Biol. Chem., 1996, 271 (18): 10866-10873. |
| 17 | Du H X, Xiao W H, Wang Y, et al. Engineering Yarrowia lipolytica for campesterol overproduction[J]. PLoS One, 2016, 11 (1): e0146773. |
| 18 | Zhang Y, Wang Y, Yao M, et al. Improved campesterol production in engineered Yarrowia lipolytica strains[J]. Biotechnol. Lett., 2017, 39 (7): 1033-1039. |
| 19 | Wong J, d'Espaux L, Dev I, et al. De novo synthesis of the sedative valerenic acid in Saccharomyces cerevisiae[J]. Metab. Eng., 2018, 47: 94-101. |
| 20 | Reider Apel A, d'Espaux L,Wehrs M, et al. A Cas9-based toolkit to program gene expression in Saccharomyces cerevisiae[J]. Nucleic Acids Res., 2017, 45 (1): 496-508. |
| 21 | Li X, Roberti R, Blobel G. Structure of an integral membrane sterol reductase from Methylomicrobium alcaliphilum[J]. Nature, 2015, 517 (7532): 104-107. |
| 22 | Witsch-Baumgartner M, Löffler J, Utermann G. Mutations in thehuman DHCR7 gene[J]. Hum. Mutat., 2001, 17 (3): 172-182. |
| 23 | Prabhu A V, Luu W, Li D, et al. DHCR7: a vital enzyme switch between cholesterol and vitamin D production[J]. Prog. Lipid Res., 2016, 64: 138-151. |
| 24 | Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments[J]. Nucleic Acids Res., 2019, 47 (W1): W256-W259. |
| 25 | Li T, Liu G S, Zhou W, et al. Metabolic engineering of Saccharomyces cerevisiae to overproduce squalene[J]. J. Agric. Food Chem., 2020, 68 (7): 2132-2138. |
| 26 | Liu G S,Li T, Zhou W, et al. The yeast peroxisome: a dynamic storage depot and subcellular factory for squalene overproduction[J]. Metab. Eng., 2020, 57: 151-161. |
| 27 | Hubmann G, Thevelein J M, Nevoigt E. Natural and modified promoters for tailored metabolic engineering of the yeast Saccharomyces cerevisiae[J]. Methods Mol. Biol., 2014, 1152: 17-42. |
| 28 | Sun J, Shao Z, Zhao H, et al. Cloning and characterization of a panel of constitutive promoters for applications in pathway engineering in Saccharomyces cerevisiae[J]. Biotechnol. Bioeng., 2012, 109 (8): 2082-2092. |
| 29 | Monfort A, Finger S, Sanz P, et al. Evaluation of different promoters for the efficient production of heterologous proteins in baker's yeast[J]. Biotechnology Letters, 1999, 21 (3): 225-229. |
| 30 | Partow S, Siewers V, Bjørn S, et al. Characterization of different promoters for designing a new expression vector in Saccharomyces cerevisiae[J]. Yeast, 2010, 27 (11): 955-964. |
| 31 | Bhattacharya S, Esquivel B D, White T C. Overexpression or deletion of ergosterol biosynthesis genes alters doubling time, response to stress agents, and drug susceptibility in Saccharomyces cerevisiae[J]. mBio, 2018, 9 (4): e01291-18. |
| 32 | Liu J F, Xia J J, Nie K L, et al. Outline of the biosynthesis and regulation of ergosterol in yeast[J]. World J. Microbiol. Biotechnol., 2019, 35(7): 98. |
| 33 | Veen M, Stahl U, Lang C. Combined overexpression of genes of the ergosterol biosynthetic pathway leads to accumulation of sterols in Saccharomyces cerevisiae[J]. FEMS Yeast Research, 2003, 4 (1): 87-95. |
| 34 | Ma B X, Ke X, Tang X L, et al. Rate-limiting steps in the Saccharomyces cerevisiae ergosterol pathway: towards improved ergosta-5,7-dien-3β-ol accumulation by metabolic engineering[J]. World J. Microbiol. Biotechnol., 2018, 34 (4): 55. |
| 35 | 张振颖, 何秀萍, 李巍巍, 等. 甾醇C-24甲基转移酶和甾醇C-8异构酶在酿酒酵母麦角甾醇生物合成中的调控作用[J]. 微生物学报, 2009, 49 (8): 1063-1068. |
| Zhang Z Y, He X P, Li W W, et al. Regulation role of sterol C-24 methyltransferase and sterol C-8 isomerase in the ergosterol biosynthesis of Saccharomyces cerevisiae[J]. Acta Microbiologica Sinica, 2009, 49 (8): 1063-1068. |
| [1] | 刘昕, 戈钧, 李春. 光驱动微生物杂合系统提高生物制造水平[J]. 化工学报, 2023, 74(1): 330-341. |
| [2] | 刘雪, 张莉娟, 赵广荣. 大肠杆菌偏利共培养系统合成大豆苷元[J]. 化工学报, 2022, 73(9): 4015-4024. |
| [3] | 孙怡, 张腾, 吕波, 李春. 胞内生物传感器提高微生物细胞工厂的精细调控[J]. 化工学报, 2022, 73(2): 521-534. |
| [4] | 王靖楠, 庞建, 秦磊, 郭超, 吕波, 李春, 王超. 丁烯基多杀菌素高产菌株的选育和改造策略[J]. 化工学报, 2022, 73(2): 566-576. |
| [5] | 王欣慧, 王颖, 姚明东, 肖文海. 维生素A生物合成的研究进展[J]. 化工学报, 2022, 73(10): 4311-4323. |
| [6] | 王欣, 赵鹏, 李清扬, 田平芳. 半导体合成生物学的研究进展[J]. 化工学报, 2021, 72(5): 2426-2435. |
| [7] | 毛金竹, 肖淑玲, 杨智淳, 王孝宇, 张诗, 陈俊宏, 谢佶晟, 陈福德, 黄子诺, 冯天宇, 张瑷珲, 方柏山. 合成生物学在农残检测领域的应用[J]. 化工学报, 2021, 72(5): 2413-2425. |
| [8] | 赵贞尧, 张保财, 李锋, 宋浩. 产电细胞的合成生物学设计构建[J]. 化工学报, 2021, 72(1): 468-482. |
| [9] | 王炼, 吴迪, 周景文. 木脂素的生物合成及其微生物法生产的研究进展[J]. 化工学报, 2021, 72(1): 320-333. |
| [10] | 王凯峰, 王金鹏, 韦萍, 纪晓俊. 代谢工程改造解脂耶氏酵母生产脂肪酸及其衍生物[J]. 化工学报, 2021, 72(1): 351-365. |
| [11] | 高虎涛, 申晓林, 孙新晓, 王佳, 袁其朋. 代谢工程调控策略在生物合成氨基酸及其衍生物中的应用[J]. 化工学报, 2020, 71(9): 4058-4070. |
| [12] | 徐静, 由紫暄, 张君奇, 陈正, 吴德光, 李锋, 宋浩. 合成生物学方法改造电活性生物膜研究进展[J]. 化工学报, 2020, 71(9): 3950-3962. |
| [13] | 秦磊, 俞杰, 宁小钰, 孙文涛, 李春. 合成生物系统构建与绿色生物“智”造[J]. 化工学报, 2020, 71(9): 3979-3994. |
| [14] | 徐彦芹, 杨锡智, 罗若诗, 黄玉红, 霍锋, 王丹. 合成生物学在生物基塑料制造中的应用[J]. 化工学报, 2020, 71(10): 4520-4531. |
| [15] | 李诺楠, 李春. 糖基转移酶在三萜皂苷合成中的应用[J]. 化工学报, 2019, 70(10): 3869-3879. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备 11010102001995号
京公网安备 11010102001995号