CIESC Journal ›› 2020, Vol. 71 ›› Issue (9): 3979-3994.DOI: 10.11949/0438-1157.20200516
• Reviews and monographs • Previous Articles Next Articles
Lei QIN1( ),Jie YU1,Xiaoyu NING1,Wentao SUN1,Chun LI1,2(
),Jie YU1,Xiaoyu NING1,Wentao SUN1,Chun LI1,2( )
)
Received:2020-05-08
Revised:2020-06-27
Online:2020-09-05
Published:2020-09-05
Contact:
Chun LI
通讯作者:
李春
作者简介:秦磊(1987—),男,博士,基金资助:CLC Number:
Lei QIN, Jie YU, Xiaoyu NING, Wentao SUN, Chun LI. Synthetic biological system construction and green intelligent biological manufacturing[J]. CIESC Journal, 2020, 71(9): 3979-3994.
秦磊, 俞杰, 宁小钰, 孙文涛, 李春. 合成生物系统构建与绿色生物“智”造[J]. 化工学报, 2020, 71(9): 3979-3994.
Add to citation manager EndNote|Ris|BibTeX

Fig.3 Intelligence of enzymes(a) synthesis of various products by enzyme promiscuity[35]; (b) changing properties of enzymes by regulating temperature[36]

Fig.4 Transcription regulatory elements as biosensors(a) rational design of IPP-response element[53]; (b) cold-induced expression system[59]; (c) light-induced and light-repressed expression system inE. coli[64]; (d) light-induced expression system in S. cerevisiae[65]
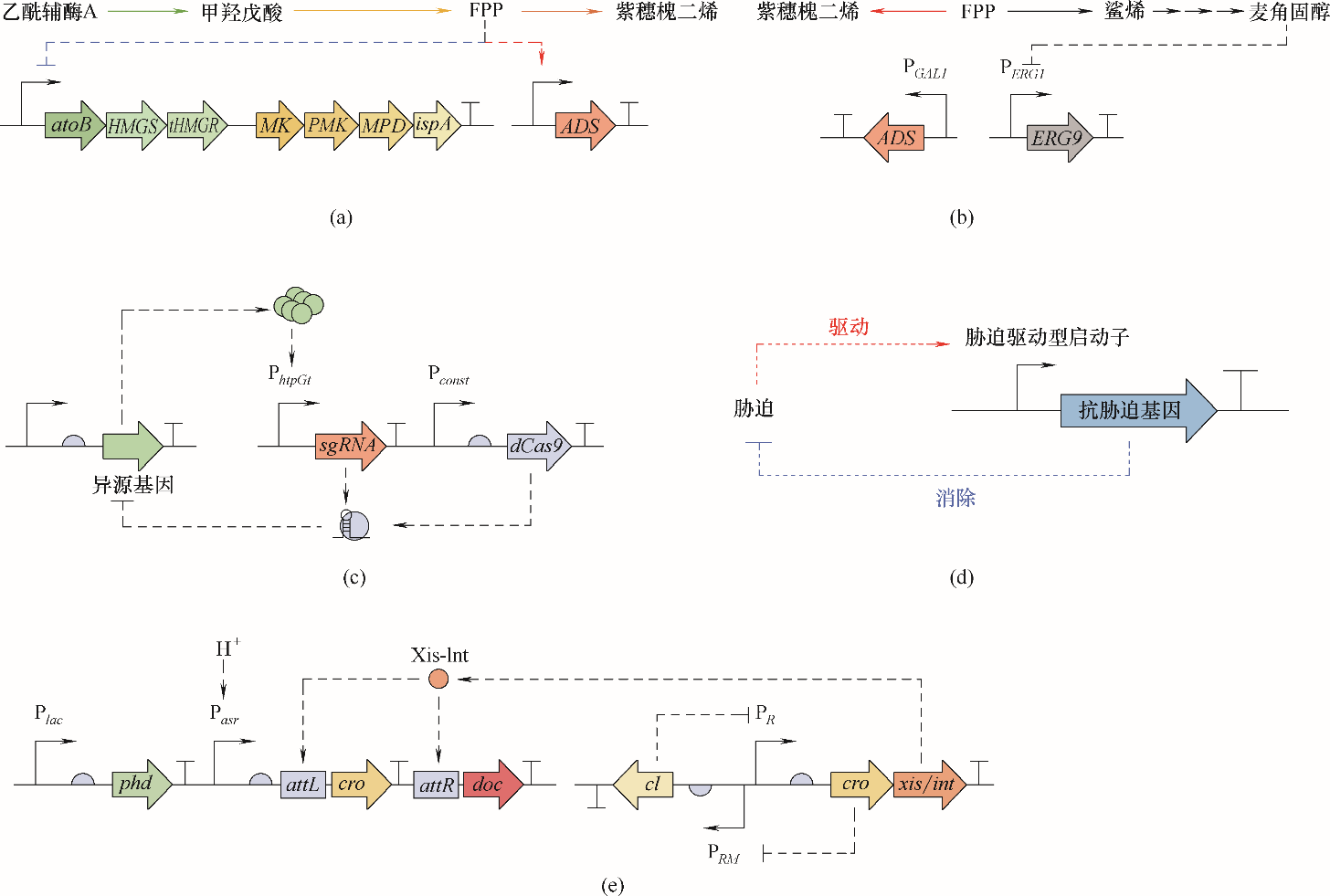
Fig.5 Autonomous dynamic regulations(a) FPP feedback inhibition and induction increase amorphadiene production[56]; (b) ergosterol feedback inhibition decreases competitive pathway flux[91]; (c) burden induced CRISPRi feedback regulation[92]; (d) stress-driven feedback regulation of anti-stress system[57]; (e) two count pH sensitive kill switch[93]
| 1 | Dai Z, Nielsen J. Advancing metabolic engineering through systems biology of industrial microorganisms[J]. Current Opinion in Biotechnology, 2015, 36: 8-15. |
| 2 | Lee J W, Na D, Park J M, et al. Systems metabolic engineering of microorganisms for natural and non-natural chemicals[J]. Nature Chemical Biology, 2012, 8(6): 536. |
| 3 | Curran K A, Alper H S. Expanding the chemical palate of cells by combining systems biology and metabolic engineering[J]. Metabolic Engineering, 2012, 14(4): 289-297. |
| 4 | Zhang F, Rodriguez S, Keasling J D. Metabolic engineering of microbial pathways for advanced biofuels production[J]. Current Opinion in Biotechnology, 2011, 22(6): 775-783. |
| 5 | Mao X, Liu Z, Sun J, et al. Metabolic engineering for the microbial production of marine bioactive compounds[J]. Biotechnology Advances, 2017, 35(8): 1004-1021. |
| 6 | Pontrelli S, Chiu T Y, Lan E I, et al. Escherichia coli as a host for metabolic engineering[J]. Metabolic Engineering, 2018, 50: 16-46. |
| 7 | Matsumoto T, Tanaka T, Kondo A. Engineering metabolic pathways in Escherichia coli for constructing a “microbial chassis” for biochemical production[J]. Bioresource Technology, 2017, 245: 1362-1368. |
| 8 | Heider S A E, Wendisch V F. Engineering microbial cell factories: metabolic engineering of Corynebacterium glutamicum with a focus on non-natural products[J]. Biotechnology Journal, 2015, 10(8): 1170-1184. |
| 9 | Becker J, Wittmann C. Bio-based production of chemicals, materials and fuels—Corynebacterium glutamicum as versatile cell factory[J]. Current Opinion in Biotechnology, 2012, 23(4): 631-640. |
| 10 | Zhang Y, Nielsen J, Liu Z. Engineering yeast metabolism for production of terpenoids for use as perfume ingredients, pharmaceuticals and biofuels[J]. FEMS Yeast Research, 2017, 17: 8. |
| 11 | Bond C M, Tang Y. Engineering Saccharomyces cerevisiae for production of simvastatin[J]. Metabolic Engineering, 2019, 51: 1-8. |
| 12 | Yaguchi A, Spagnuolo M, Blenner M. Engineering yeast for utilization of alternative feedstocks[J]. Current Opinion in Biotechnology, 2018, 53: 122-129. |
| 13 | Ko J K, Lee S M. Advances in cellulosic conversion to fuels: engineering yeasts for cellulosic bioethanol and biodiesel production[J]. Current Opinion in Biotechnology, 2018, 50: 72-80. |
| 14 | Zhu M, Wang C, Sun W, et al. Boosting 11-oxo-β-amyrin and glycyrrhetinic acid synthesis in Saccharomyces cerevisiaevia pairing novel oxidation and reduction system from legume plants[J]. Metabolic Engineering, 2018, 45: 43-50. |
| 15 | Biggs B W, De Paepe B, Santos C N S, et al. Multivariate modular metabolic engineering for pathway and strain optimization[J]. Current Opinion in Biotechnology, 2014, 29: 156-162. |
| 16 | Caspeta L, Castillo T, Nielsen J. Modifying yeast tolerance to inhibitory conditions of ethanol production processes[J]. Frontiers in Bioengineering and Biotechnology, 2015, 3: 184. |
| 17 | Cunha J T, Romaní A, Costa C E, et al. Molecular and physiological basis of Saccharomyces cerevisiae tolerance to adverse lignocellulose-based process conditions[J]. Applied Microbiology and Biotechnology, 2019, 103: 159-175. |
| 18 | Xu K, Lv B, Huo Y X, et al. Toward the lowest energy consumption and emission in biofuel production: combination of ideal reactors and robust hosts[J]. Current Opinion in Biotechnology, 2017, 50: 19-24. |
| 19 | Deparis Q, Claes A, Foulquié-Moreno M R, et al. Engineering tolerance to industrially relevant stress factors in yeast cell factories[J]. FEMS Yeast Research, 2017, 17: 4. |
| 20 | Morano K A, Grant C M, Moye-Rowley W S. The response to heat shock and oxidative stress in Saccharomyces cerevisiae[J]. Genetics, 2012, 190(4): 1157-1195. |
| 21 | Xu K, Lee Y S, Li J, et al. Resistance mechanisms and reprogramming of microorganisms for efficient biorefinery under multiple environmental stresses[J]. Synthetic and Systems Biotechnology, 2019, 4(2): 92-98. |
| 22 | Xu K, Qin L, Bai W, et al. Multilevel defense system (MDS) relieves multiple stresses for economically boosting ethanol production of industrial Saccharomyces cerevisiae[J]. ACS Energy Letters, 2020, 5(2): 572-582. |
| 23 | Leman J K, Weitzner B D, Lewis S M, et al. Macromolecular modeling and design in Rosetta: recent methods and frameworks[J]. Nature Methods, 2020, 17: 665-680. |
| 24 | Weitzner B D, Kipnis Y, Daniel A G, et al. A computational method for design of connected catalytic networks in proteins[J]. Protein Science, 2019, 28(12): 2036-2041. |
| 25 | Chen Z, Boyken S E, Jia M, et al. Programmable design of orthogonal protein heterodimers[J]. Nature, 2019, 565: 106-111. |
| 26 | Boyken S E, Benhaim M A, Busch F, et al. De novo design of tunable, pH-driven conformational changes[J]. Science, 2019, 364: 658-664. |
| 27 | Zhang Y, Bartz R. Grigoryan G,et al. Computational design and experimental characterization of peptides intended for pH-dependent membrane insertion and pore formation[J]. ACS Chemical Biology, 2015, 10(4): 1082-1093. |
| 28 | Kisovec M, Rezelj S, Knap P, et al. Engineering a pH responsive pore forming protein[J]. Scientific Reports, 2017, 7: 42231. |
| 29 | Omersa N, Aden S, Kisovec M, et al. Design of protein logic gate system operating on lipid membranes[J]. ACS Synthetic Biology, 2020, 9: 316-328. |
| 30 | Langan R A, Boyken S E, Ng A H, et al. De novo design of bioactive protein switches[J]. Nature, 2019, 572: 205-210. |
| 31 | Ng A H, Nguyen T H, Gómez-Schiavon M, et al. Modular and tunable biological feedback control using a de novo protein switch[J]. Nature, 2019, 572: 265-269. |
| 32 | Chen Z, Kibler R D, Hunt A, et al. De novo design of protein logic gates[J]. Science, 2020, 368: 78-84. |
| 33 | Tinberg C E, Khare S D, Dou J, et al. Computational design of ligand-binding proteins with high affinity and selectivity[J]. Nature, 2013, 501(7466): 212-216. |
| 34 | Urlacher V B, Girhard M. Cytochrome P450 monooxygenases in biotechnology and synthetic biology[J]. Trends in Biotechnology, 2019, 37: 882. |
| 35 | Sun W, Xue H, Liu H, et al. Controlling chemo- and regioselectivity of a plant P450 in yeast cell toward rare licorice triterpenoid biosynthesis[J]. ACS Catalysis, 2020, 10: 4253-4260. |
| 36 | Inda M E, Vandenbranden M, Fernández A, et al. A lipid-mediated conformational switch modulates the thermosensing activity of DesK[J]. Proceedings of the National Academy of Sciences of the USA, 2014, 111(9): 3579-3584. |
| 37 | Wu M Y, Sung L Y, Li H, et al. Combining CRISPR and CRISPRi systems for metabolic engineering of E. coli and 1,4-BDO biosynthesis[J]. ACS Synthetic Biology, 2017, 6: 2350-2361. |
| 38 | Rodrigues A L, Becker J, de Souza Lima A O, et al. Systems metabolic engineering of Escherichia coli for gram scale production of the antitumor drug deoxyviolacein from glycerol[J]. Biotechnology and Bioengineering, 2014, 111: 2280-2289. |
| 39 | Wang S, Hou Y, Chen X, et al. Kick-starting evolution efficiency with an autonomous evolution mutation system[J]. Metabolic Engineering, 2019, 54: 127-136. |
| 40 | Xie W, Ye L, Lv X, et al. Sequential control of biosynthetic pathways for balanced utilization of metabolic intermediates in Saccharomyces cerevisiae[J]. Metabolic Engineering, 2015, 28: 8-18. |
| 41 | Paddon C J, Westfall P J, Pitera D J, et al. High-level semi-synthetic production of the potent antimalarial artemisinin[J]. Nature, 2013, 496: 528-532. |
| 42 | Šeputiene V, Motiejūnas D, Suziedelis K, et al. Molecular characterization of the acid-inducible asr gene of Escherichia coli and its role in acid stress response[J]. Journal of Bacteriology, 2003, 185(8): 2475-2484. |
| 43 | Ogasawara H, Hasegawa A, Kanda E, et al. Genomic SELEX search for target promoters under the control of the PhoQP-RstBA signal relay cascade[J]. Journal of Bacteriology, 2007, 189(13): 4791-4799. |
| 44 | Hoynes-O’Connor A, Shopera T, Hinman K, et al. Enabling complex genetic circuits to respond to extrinsic environmental signals[J]. Biotechnology and Bioengineering, 2017, 114: 1626-1631. |
| 45 | Haneburger I, Fritz G, Jurkschat N, et al. Deactivation of the E. coli pH stress sensor CadC by cadaverine[J]. Journal of Molecular Biology, 2012, 424: 15-27. |
| 46 | Moser F, Borujeni A E, Ghodasara A N, et al. Dynamic control of endogenous metabolism with combinatorial logic circuits[J]. Molecular Systems Biology, 2018, 14: 8605. |
| 47 | Xiong L, Zeng Y, Tang R Q, et al. Condition specific promoter activities in Saccharomyces cerevisiae[J]. Microbial Cell Factories, 2018, 17: 58. |
| 48 | Rajkumar A S, Liu G d, Bergenholm D, et al. Engineering of synthetic, stress-responsive yeast promoters[J]. Nucleic Acids Research, 2016, 44(17): 136. |
| 49 | Hanko E K R, Paiva A C, Jonczyk M, et al. A genome-wide approach for identification and characterisation of metabolite-inducible systems[J]. Nature Communications, 2020, 11: 1213. |
| 50 | Xu P, Wang W, Li L, et al. Design and kinetic analysis of a hybrid promoter-regulator system for malonyl-CoA sensing in Escherichia coli[J]. ACS Chemical Biology, 2014, 9(2): 451-458. |
| 51 | Xu P, Li L, Zhang F, et al. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control[J]. Proceedings of the National Academy of Sciences of the USA, 2014, 111(31): 11299-11304. |
| 52 | Liang C, Zhang X, Wu J, et al. Dynamic control of toxic natural product biosynthesis by an artificial regulatory circuit[J]. Metabolic Engineering, 2020, 57: 239-246. |
| 53 | Chou H H, Keasling J D. Programming adaptive control to evolve increased metabolite production[J]. Nature Communications, 2013, 4: 2595. |
| 54 | Feng J, Jester B W, Tinberg C E, et al. A general strategy to construct small molecule biosensors in eukaryotes[J]. Elife, 2015, 4: e10606. |
| 55 | Jester B W, Tinberg C E, Rich M S, et al. Engineered biosensors from dimeric ligand-binding domains[J]. ACS Synthetic Biology, 2018, 7: 2457-2467. |
| 56 | Dahl R H, Zhang F, Alonso-Gutierrez J, et al. Engineering dynamic pathway regulation using stress-response promoters[J]. Nature Biotechnology, 2013, 31(11): 1039-1046. |
| 57 | Qin L, Dong S, Yu J, et al. Stress-driven dynamic regulation of multiple tolerance genes improves robustness and productive capacity of Saccharomyces cerevisiae in industrial lignocellulose fermentation[J]. Metabolic Engineering, 2020, 61:160-170. |
| 58 | Valdez-Cruz N A, Caspeta L, Pérez N O, et al. Production of recombinant proteins in E. coli by the heat inducible expression system based on the phage lambda pL and/or pR promoters[J]. Microbial Cell Factories, 2010, 9: 18. |
| 59 | Zheng Y, Meng F, Zhu Z, et al. A tight cold-inducible switch built by coupling thermosensitive transcriptional and proteolytic regulatory parts[J]. Nucleic Acids Research, 2019, 47(21): 137. |
| 60 | Guan C R, Cui W J, Cheng J T, et al. Construction and development of an auto-regulatory gene expression system in Bacillus subtilis[J]. Microbial Cell Factories, 2015, 14: 150. |
| 61 | Fuqua C, Parsek M R, Greenberg E P. Regulation of gene expression by cell-to-cell communication: acylhomoserine lactone quorum sensing[J]. Annual Review of Genetics, 2001, 35: 439-468. |
| 62 | Jia H, Sun X, Sun H, et al. Intelligent microbial heat-regulating engine (IMHeRE) for improved thermo-robustness and efficiency of bioconversion[J]. ACS Synthetic Biology, 2016, 5: 312-320. |
| 63 | Zoltowski B D, Motta-Mena L B, Gardner K H. Blue light-induced dimerization of a bacterial LOV-HTH DNA-binding protein[J]. Biochemistry, 2013, 52: 6653-6661. |
| 64 | Jayaraman P, Devarajan K, Chua T K, et al. Blue light-mediated transcriptional activation and repression of gene expression in bacteria[J]. Nucleic Acids Research, 2016, 44: 6994-7005. |
| 65 | Zhao E M, Zhang Y, Mehl J, et al. Optogenetic regulation of engineered cellular metabolism for microbial chemical production[J]. Nature, 2018, 555: 683. |
| 66 | Hochrein L, Machens F, Messerschmidt K, et al. PhiReX: a programmable and red light-regulated protein expression switch for yeast[J]. Nucleic Acids Research, 2017, 110: 21130-21135. |
| 67 | Schmidl S R, Sheth R U, Wu A, et al. Refactoring and optimization of light-switchable Escherichia coli two component systems[J]. ACS Synthetic Biology, 2014, 3: 820-831. |
| 68 | Olson E J, Hartsough L A, Landry B P, et al. Characterizing bacterial gene circuit dynamics with optically programmed gene expression signals[J]. Nature Methods, 2014, 11: 449-455. |
| 69 | Neupert J, Karcher D, Bock R. Design of simple synthetic RNA thermometers for temperature-controlled gene expression in Escherichia coli[J]. Nucleic Acids Research, 2008, 36: 124. |
| 70 | Giuliodori A M, Di Pietro F, Marzi S, et al. The cspA mRNA is a thermosensor that modulates translation of the cold-shock protein CspA[J]. Molecular Cell, 2010, 37(1): 21-33. |
| 71 | Nechooshtan G, Elgrably-Weiss M, Sheaffer A, et al. A pH-responsive riboregulatory[J]. Genes and Development, 2009, 23: 2650-2662. |
| 72 | Nechooshtan G, Elgrably-Weiss M, Altuvia S. Changes in transcriptional pausing modify the folding dynamics of the pH-responsive RNA element[J]. Nucleic Acids Research, 2014, 42: 622-630. |
| 73 | Pham H L, Wong A, Chua N, et al. Engineering a riboswitch-based genetic platform for the self-directed evolution of acid-tolerant phenotypes[J]. Nature Communications, 2017, 8: 411. |
| 74 | Lukyanov K A, Belousov V V. Genetically encoded fluorescent redox sensors[J]. Biochimica et Biophysica Acta, 2014, 1840(2): 745-756. |
| 75 | Bilan D S, Belousov V V. New tools for redox biology: from imaging to manipulation[J]. Free Radical Biology and Medicine, 2017, 109: 167-188. |
| 76 | 俞杰, 秦磊, 许可, 等. 细胞工厂氧化还原状态的荧光探针检测与调控[J].生物加工过程, 2020, 18(1): 60-69. |
| Yu J, Qin L, Xu K, et al. Detection and regulation of the redox state in cell factories by fluorescent probes[J]. Chinese Journal of Bioprocess Engineering, 2020, 18(1): 60-69. | |
| 77 | Bugaj L J, Choksi A T, Mesuda C K, et al. Optogenetic protein clustering and signaling activation in mammalian cells[J]. Nature Methods, 2013, 10: 249-252. |
| 78 | Taslimi A, Vrana J D, Chen D, et al. An optimized optogenetic clustering tool for probing protein interaction and function[J]. Nature Communications, 2014, 5: 4925. |
| 79 | Shin Y, Berry J, Pannucci N, et al. Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets[J]. Cell, 2017, 168: 159-171. |
| 80 | Zhao E M, Suek N, Wilson M Z, et al. Light-based control of metabolic flux through assembly of synthetic organelles[J]. Nature Chemical Biology, 2019, 15: 589-597. |
| 81 | Dine E, Gil A A, Uribe G, et al. Protein phase separation provides long-term memory of transient spatial stimuli[J]. Cell Systems, 2018, 6: 655-663. |
| 82 | Gil A A, Laptenok S P, Iuliano J N, et al. Photoactivation of the BLUF protein PixD probed by the site-specific incorporation of fluorotyrosine residues[J]. Journal of the American Chemical Society, 2017, 139: 14638-14648. |
| 83 | Kim S K, Han G H, Seong W, et al. CRISPR interference-guided balancing of a biosynthetic mevalonate pathway increases terpenoid production[J]. Metabolic Engineering, 2016, 38: 228-240. |
| 84 | Cunningham-Bryant D, Sun J, Fernandez B, et al. CRISPR-Cas-mediated chemical control of transcriptional dynamics in yeast[J]. ChemBioChem, 2019, 20(12): 1519-1523. |
| 85 | Koopal B, Kruis A J, Claassens N J, et al. Incorporation of a synthetic amino acid into dCas9 improves control of gene silencing[J]. ACS Synthetic Biology, 2019, 8(2): 216-222. |
| 86 | Ni J, Wu Y T, Tao F, et al. A coenzyme-free biocatalyst for the value-added utilization of lignin-derived aromatics[J]. Journal of the American Chemical Society, 2018, 140: 16001-16005. |
| 87 | Bañares A B, Valdehuesa K N G, Ramos K R M, et al. A pH-responsive genetic sensor for the dynamic regulation of D-xylonic acid accumulation in Escherichia coli[J]. Applied Microbiology and Biotechnology, 2020, 104: 2097-2108. |
| 88 | Bañares A B, Valdehuesa K N G, Ramos K R M, et al. Discovering a novel D-xylonate-responsive promoter: the PyjhI-driven genetic switch towards better 1,2,4-butanetriol production[J]. Applied Microbiology and Biotechnology, 2019, 103: 8063-8074. |
| 89 | Anesiadis N, Kobayashi H, Cluett W R, et al. Analysis and design of a genetic circuit for dynamic metabolic engineering[J]. ACS Synthetic Biology, 2013, 2(8): 442-452. |
| 90 | Kim E M, Woo H M, Tian T, et al. Autonomous control of metabolic state by a quorum sensing (QS)-mediated regulator for bisabolene production in engineered E. coli[J]. Metabolic Engineering, 2017, 44: 325-336. |
| 91 | Yuan J, Ching C B. Dynamic control of ERG9 expression for improved amorpha-4,11-diene production in Saccharomyces cerevisiae[J]. Microbial Cell Factories, 2015, 14: 38. |
| 92 | Ceroni F, Boo A, Furini S, et al. Burden-driven feedback control of gene expression[J]. Nature Methods, 2018, 15(5): 387-393. |
| 93 | Stirling F, Naydich A, Bramante J, et al. Synthetic cassettes for pH-mediated sensing, counting and containment[J]. Cell Reports, 2020, 30(9): 3139. |
| 94 | Zhao N, Bai Y, Liu C G, et al. Flocculating Zymomonas mobilis is a promising host to be engineered for fuel ethanol production from lignocellulosic biomass[J]. Biotechnology Journal, 2014, 9(3): 362-371. |
| 95 | Govender P, Domingo J L, Bester M C, et al. Controlled expression of the dominant flocculation genes FLO1, FLO5, and FLO11 in Saccharomyces cerevisiae[J]. Applied and Environmental Microbiology, 2008, 74(19): 6041-6052. |
| 96 | Li Q, Zhao X Q, Chang A K, et al. Ethanol-induced yeast flocculation directed by the promoter of TPS1 encoding trehalose-6-phosphate synthase 1 for efficient ethanol production[J]. Metabolic Engineering, 2012, 14: 1-8. |
| 97 | Ling C, Qiao G Q, Shuai B W, et al. Engineering self-flocculating Halomonas campaniensis for wastewaterless open and continuous fermentation[J]. Biotechnology and Bioengineering, 2019, 116: 805-815. |
| 98 | Heler R, Wright A V, Vucelja M, et al. Mutations in Cas9 enhance the rate of acquisition of viral spacer sequences during the CRISPR-Cas immune response[J]. Molecular Cell, 2017, 65(1): 168-175. |
| 99 | Jiang W, Oikonomou P, Tavazoie S. Comprehensive genome-wide perturbations via CRISPR adaptation reveal complex genetics of antibiotic sensitivity[J]. Cell, 2020, 180: 1-16. |
| 100 | Xie Z X, Li B Z, Mitchell L A, et al. ‘Perfect’ designer chromosome V and behavior of a ring derivative[J]. Science, 2017, 355(6329): 4704. |
| 101 | Wu Y, Li B Z, Zhao M, et al. Bug mapping and fitness testing of chemically synthesized chromosome X[J]. Science, 2017, 355(6329): 4706. |
| 102 | Dymond J S, Richardson S M, Coombes C E, et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design[J]. Nature, 2011, 477: 471-476. |
| 103 | Jia B, Wu Y, Li B Z, et al. Precise control of SCRaMbLE in synthetic haploid and diploid yeast[J]. Nature Communications, 2018, 9(1): 1933. |
| 104 | Hochrein L, Mitchell L A, Schulz K, et al. L-SCRaMbLE as a tool for light-controlled Cre-mediated recombination in yeast[J]. Nature Communications, 2018, 9(1): 1931. |
| 105 | Ma L, Li Y, Chen X, et al. SCRaMbLE generates evolved yeasts with increased alkali tolerance[J]. Microbial Cell Factories, 2019, 18(1): 52. |
| 106 | Shen M J, Wu Y, Yang K, et al. Heterozygous diploid and interspecies SCRaMbLEing[J]. Nature Communications, 2018, 9(1): 1934. |
| 107 | Luo Z, Wang L, Wang Y, et al. Identifying and characterizing SCRaMbLEd synthetic yeast using ReSCuES[J]. Nature Communications, 2018, 9(1): 1930. |
| 108 | Blount B A, Gowers G O F, Ho J C H, et al. Rapid host strain improvement by in vivo rearrangement of a synthetic yeast chromosome[J]. Nature Communications, 2018, 9(1): 1932. |
| 109 | Wu Y, Zhu R Y, Mitchell L A, et al. In vitro DNA SCRaMbLE[J]. Nature Communications, 2018, 9(1): 1935. |
| 110 | Wang J, Xie Z X, Ma Y, et al. Ring synthetic chromosome V SCRaMbLE[J]. Nature Communications, 2018, 9(1): 3783. |
| 111 | Gowers G O F, Chee S M, Bell D, et al. Improved betulinic acid biosynthesis using synthetic yeast chromosome recombination and semi-automated rapid LC-MS screening[J]. Nature Communications, 2020, 11(1): 868. |
| 112 | Liu W, Luo Z, Wang Y, et al. Rapid pathway prototyping and engineering using in vitro and in vivo synthetic genome SCRaMbLE-in methods[J]. Nature Communications, 2018, 9(1): 1936. |
| 113 | 田锡炜, 王冠, 张嗣良, 等. 工业生物过程智能控制原理和方法进展[J]. 生物工程学报, 2019, 35(10): 2014-2024. |
| Tian X W, Wang G, Zhang S L, et al. Progress in intelligent control of industrial bioprocess[J]. Chinese Journal of Biotechnology, 2019, 35(10): 2014-2024. | |
| 114 | Chen Y, Wang Z J, Chu J, et al. Significant decrease of broth viscosity and glucose consumption in erythromycin fermentation by dynamic regulation of ammonium sulfate and phosphate[J]. Bioresource Technology, 2013, 134: 173-179. |
| 115 | Zou X, Xia J Y, Chu J, et al. Real-time fluid dynamics investigation and physiological response for erythromycin fermentation scale-up from 50 L to 132 m3 fermenter[J]. Bioprocess and Biosystems Engineering, 2012, 35(5): 789-800. |
| 116 | 陈晓春. 啤酒发酵系统温度智能控制[J]. 食品工业, 2019, 40(11): 219-221. |
| Chen X C. Intelligent temperature control of beer fermentation system[J]. The Food Industry, 2019, 40(11): 219-221. | |
| 117 | Wang L, Yuan J, Wu C, et al. Practical algorithm for stochastic optimal control problem about microbial fermentation in batch culture[J]. Optimization Letters, 2019, 13: 527-541. |
| 118 | Grunberger A, Wiechert W, Kohlheyer D. Single-cell microfluidics: opportunity for bioprocess development[J]. Current Opinion in Biotechnology, 2014, 29: 15-23. |
| 119 | Chen J, Vestergaard M, Shen J, et al. Droplet-based microfluidics as a future tool for strain improvement in lactic acid bacteria[J]. FEMS Microbiology Letters, 2018, 365: 258. |
| 120 | Macosko E Z, Basu A, Satija R, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets[J]. Cell, 2015, 161: 1202-1214. |
| 121 | Blattman S B, Jiang W, Oikonomou P, et al. Prokaryotic single-cell RNA sequencing by in situ combinatorial indexing[J]. Nature Microbiology, 2020, . |
| 122 | Si T, Chao R, Min Y, et al. Automated multiplex genome-scale engineering in yeast[J]. Nature Communications, 2017, 8: 15187. |
| [1] | Ruiqi LIU, Xitong ZHOU, Yue ZHANG, Ying HE, Jing GAO, Li MA. The construction and application of biosensor based on gold nanoparticles loaded SiO2-nanoflowers [J]. CIESC Journal, 2023, 74(3): 1247-1259. |
| [2] | Xin LIU, Jun GE, Chun LI. Light-driven microbial hybrid systems improve level of biomanufacturing [J]. CIESC Journal, 2023, 74(1): 330-341. |
| [3] | Tianhang ZHOU, Xingying LAN, Chunming XU. Artificial intelligence for accelerating polymer design: recent advances and future perspectives [J]. CIESC Journal, 2023, 74(1): 14-28. |
| [4] | Xue LIU, Lijuan ZHANG, Guangrong ZHAO. Commensalistic Escherichia coli coculture for biosynthesis of daidzein [J]. CIESC Journal, 2022, 73(9): 4015-4024. |
| [5] | Jingnan WANG, Jian PANG, Lei QIN, Chao GUO, Bo LYU, Chun LI, Chao WANG. Breeding and modification strategies of butenyl-spinosyn high-yield strains [J]. CIESC Journal, 2022, 73(2): 566-576. |
| [6] | Yi SUN, Teng ZHANG, Bo LYU, Chun LI. Improvement for fine regulation of microbial cell factory by intracellular biosensors [J]. CIESC Journal, 2022, 73(2): 521-534. |
| [7] | Xinhui WANG, Ying WANG, Mingdong YAO, Wenhai XIAO. Research progress of vitamin A biosynthesis [J]. CIESC Journal, 2022, 73(10): 4311-4323. |
| [8] | Wulin ZHOU, Huifang GAO, Yuling WU, Xian ZHANG, Meijuan XU, Taowei YANG, Minglong SHAO, Zhiming RAO. Engineering of Saccharomyces cerevisiae for biosynthesis of campesterol [J]. CIESC Journal, 2021, 72(8): 4314-4324. |
| [9] | WANG Yali,FU Yousi,CHEN Junhong,HUANG Jiacheng,LIAO Langxing,ZHANG Yonghui,FANG Baishan. Enzyme engineering: from artificial design to artificial intelligence [J]. CIESC Journal, 2021, 72(7): 3590-3600. |
| [10] | CHEN Tingting, HAN Kaixin, CHEN Cuixue, LING Xueping, SHEN Liang, LU Yinghua. Study of iron-reducing bacteria Shewanellaxiamenensis BC01 under organic solvents stress [J]. CIESC Journal, 2021, 72(7): 3747-3756. |
| [11] | YANG Ruixiong, ZHENG Xin, LU Tao, ZHAO Yuze, YANG Qinghua, LU Yinghua, HE Ning, LING Xueping. Effects of substitution of ER domains on the synthesis of eicosapentaenoic acid in Schizochytrium limacinum SR21 [J]. CIESC Journal, 2021, 72(7): 3768-3779. |
| [12] | MAO Jinzhu, XIAO Shuling, YANG Zhichun, WANG Xiaoyu, ZHANG Shi, CHEN Junhong, XIE Jisheng, CHEN Fude, HUANG Zinuo, FENG Tianyu, ZHANG Aihui, FANG Baishan. Application of synthetic biology in pesticides residues detection [J]. CIESC Journal, 2021, 72(5): 2413-2425. |
| [13] | WANG Xin, ZHAO Peng, LI Qingyang, TIAN Pingfang. Research advances in semiconductor synthetic biology [J]. CIESC Journal, 2021, 72(5): 2426-2435. |
| [14] | Nan SU, Yinan WU, Yinyee TAN, Lihua JIN, Chong ZHANG, Aikawa SHIMPEI, Hasunuma TOMOHISA, Kondo AKIHIKO, Xinhui XING. Comparative omics study of Spirulinaplatensis mutants based on ARTP mutagenesis breeding system [J]. CIESC Journal, 2021, 72(12): 6298-6310. |
| [15] | ZHAO Zhenyao, ZHANG Baocai, LI Feng, SONG Hao. Design and construction of exoelectrogens by synthetic biology [J]. CIESC Journal, 2021, 72(1): 468-482. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||