CIESC Journal ›› 2021, Vol. 72 ›› Issue (8): 4314-4324.DOI: 10.11949/0438-1157.20201686
• Biochemical engineering and technology • Previous Articles Next Articles
Wulin ZHOU( ),Huifang GAO,Yuling WU,Xian ZHANG,Meijuan XU,Taowei YANG,Minglong SHAO(
),Huifang GAO,Yuling WU,Xian ZHANG,Meijuan XU,Taowei YANG,Minglong SHAO( ),Zhiming RAO(
),Zhiming RAO( )
)
Received:2020-11-25
Revised:2021-01-19
Online:2021-08-05
Published:2021-08-05
Contact:
Minglong SHAO,Zhiming RAO
周武林( ),高惠芳,吴玉玲,张显,徐美娟,杨套伟,邵明龙(
),高惠芳,吴玉玲,张显,徐美娟,杨套伟,邵明龙( ),饶志明(
),饶志明( )
)
通讯作者:
邵明龙,饶志明
作者简介:周武林(1997—),男,硕士研究生, 基金资助:CLC Number:
Wulin ZHOU, Huifang GAO, Yuling WU, Xian ZHANG, Meijuan XU, Taowei YANG, Minglong SHAO, Zhiming RAO. Engineering of Saccharomyces cerevisiae for biosynthesis of campesterol[J]. CIESC Journal, 2021, 72(8): 4314-4324.
周武林, 高惠芳, 吴玉玲, 张显, 徐美娟, 杨套伟, 邵明龙, 饶志明. 重组酿酒酵母生物合成菜油甾醇[J]. 化工学报, 2021, 72(8): 4314-4324.
Add to citation manager EndNote|Ris|BibTeX
| 菌株和质粒 | 性质 | 来源 |
|---|---|---|
菌株 S.cerevisiae | ||
| GTy23 | ERG9::KanMX_PCTR3-ERG9LEU2-3, 112::His3MX6_PGAL1 ERG19/PGAL10-ERG8 URA3-52::URA3_PGAL1-mvaS (A110G)/PGAL10-mvaE(CO) HIS3Δ1::hphMX4_PGAL1ERG12/PGAL10-IDI1 | Keasling教授惠赠[ |
| Zw501 | GTy23 (ura3-52 prototrophy removed for use of Cas9 system) | 本研究构建 |
| Zw502 | Zw501 (敲除了erg5) | 本研究构建 |
| Zw503 | Zw501 (erg5::GAL1p-DrDHCR7-ADH1t) | 本研究构建 |
| Zw504 | Zw501 (erg5::GAL1p-CaDHCR7-ADH1t) | 本研究构建 |
| Zw505 | Zw501 (erg5::GAL1p-TtDHCR7-ADH1t) | 本研究构建 |
| Zw506 | Zw501 (erg5::GAL1p-CmDHCR7-ADH1t) | 本研究构建 |
| Zw507 | Zw501 (erg5::GAL1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw508 | Zw501 (erg5::GAL1p-CcDHCR7-ADH1t) | 本研究构建 |
| Zw509 | Zw501 (erg5::GAL1p-LrDHCR7-ADH1t) | 本研究构建 |
| Zw510 | Zw501 (erg5::GAL1p-CgDHCR7-ADH1t) | 本研究构建 |
| Zw511 | Zw501 (erg5::GAL1p-XmDHCR7-ADH1t) | 本研究构建 |
| Zw512 | Zw501 (erg5::GAL1p-AmDHCR7-ADH1t) | 本研究构建 |
| Zw513 | Zw501 (erg5::TPI1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw514 | Zw501 (erg5::TEF1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw515 | Zw501 (erg5::TDH3p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw516 | Zw501 (erg5::TEF2p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw517 | Zw501 (erg5::PGK1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw518 | Zw501 (erg5::GPM1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw519 | Zw501 (erg5::GPD1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw520 | Zw501 (erg5::TDH2p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw521 | Zw501 (erg5::ACT1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw522 | Zw514 (1114a::TEF1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw523 | Zw522 (607b::TEF1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw524 | Zw523 (911b::TEF1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw525 | Zw524 (1014a::TEF1p-PhDHCR7-ADH1t) | 本研究构建 |
| Escherichia coli | ||
| DH5α | 用于本研究中质粒的构建与克隆 |
Table 1 Strains and plasmids used in this study
| 菌株和质粒 | 性质 | 来源 |
|---|---|---|
菌株 S.cerevisiae | ||
| GTy23 | ERG9::KanMX_PCTR3-ERG9LEU2-3, 112::His3MX6_PGAL1 ERG19/PGAL10-ERG8 URA3-52::URA3_PGAL1-mvaS (A110G)/PGAL10-mvaE(CO) HIS3Δ1::hphMX4_PGAL1ERG12/PGAL10-IDI1 | Keasling教授惠赠[ |
| Zw501 | GTy23 (ura3-52 prototrophy removed for use of Cas9 system) | 本研究构建 |
| Zw502 | Zw501 (敲除了erg5) | 本研究构建 |
| Zw503 | Zw501 (erg5::GAL1p-DrDHCR7-ADH1t) | 本研究构建 |
| Zw504 | Zw501 (erg5::GAL1p-CaDHCR7-ADH1t) | 本研究构建 |
| Zw505 | Zw501 (erg5::GAL1p-TtDHCR7-ADH1t) | 本研究构建 |
| Zw506 | Zw501 (erg5::GAL1p-CmDHCR7-ADH1t) | 本研究构建 |
| Zw507 | Zw501 (erg5::GAL1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw508 | Zw501 (erg5::GAL1p-CcDHCR7-ADH1t) | 本研究构建 |
| Zw509 | Zw501 (erg5::GAL1p-LrDHCR7-ADH1t) | 本研究构建 |
| Zw510 | Zw501 (erg5::GAL1p-CgDHCR7-ADH1t) | 本研究构建 |
| Zw511 | Zw501 (erg5::GAL1p-XmDHCR7-ADH1t) | 本研究构建 |
| Zw512 | Zw501 (erg5::GAL1p-AmDHCR7-ADH1t) | 本研究构建 |
| Zw513 | Zw501 (erg5::TPI1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw514 | Zw501 (erg5::TEF1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw515 | Zw501 (erg5::TDH3p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw516 | Zw501 (erg5::TEF2p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw517 | Zw501 (erg5::PGK1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw518 | Zw501 (erg5::GPM1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw519 | Zw501 (erg5::GPD1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw520 | Zw501 (erg5::TDH2p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw521 | Zw501 (erg5::ACT1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw522 | Zw514 (1114a::TEF1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw523 | Zw522 (607b::TEF1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw524 | Zw523 (911b::TEF1p-PhDHCR7-ADH1t) | 本研究构建 |
| Zw525 | Zw524 (1014a::TEF1p-PhDHCR7-ADH1t) | 本研究构建 |
| Escherichia coli | ||
| DH5α | 用于本研究中质粒的构建与克隆 |
| 引物 | 序列(5′→3′) |
|---|---|
| F-up-erg5 (delet) | TGGGAATACTGTACCAGATAATCAAACAT |
| R-up-erg5 (delet) | CAAAGTTCTGTTTTTCCCCATTTGTTAAAAGGTATTTATTGTCTATTGGAATAGC |
| F-down-erg5 (delet) | ATAAATACCTTTTAACAAATGGGGAAAAACAGAACTTTGTCCAGAC |
| R-down-erg5 (delet) | TGACAGTGACGAACGCTTCAG |
| F-erg5 | ATGAGTTCTGTCGCAGAAAATATAATAC |
| R-erg5 | TTATTCGAAGACTTCTCCAGTAATTGGG |
| F-up-erg5 | TGGGAATACTGTACCAGATAATCAAACATTAAA |
| R-up-erg5 | TGTTTATACGCTATTATCAGCCAATTTGTTAAAAGGTATTTATTGTCTATTGGAATAGCA |
| F-GAL1p | CCAATAGACAATAAATACCTTTTAACAAATTGGCTGATAATAGCGTATAAACAATGCA |
| R- GAL1p | TAACTCTATCAGAAGCCATCATTTTGTAATTAAAACTTAGATTAGATTGCTATGCTTTCT |
| F-DrDHCR7 | ATCTAAGTTTTAATTACAAAATGATGGCTTCTGATAGAGTTAGAAAAAG |
| R-DrDHCR7 | ATAAATCATAAGAAATTCGCTTAAAAAATATTTGGCAACAATCTATAAGAAACAGCAG |
| F-PhDHCR7 | ATCTAAGTTTTAATTACAAAATGTCTACCTCTGAAGGTGTTAGAAAAAG |
| R-PhDHCR7 | ATAAATCATAAGAAATTCGCTTAAAAAATACCTGGCAACAATCTATATCTAACGG |
| F-LrDHCR7 | ATCTAAGTTTTAATTACAAAATGGGTAGAGTTAAATGGAGATCTATTACC |
| R-LrDHCR7 | ATAAATCATAAGAAATTCGCTTAAAAAATATTTGGCAACAATCTATATGGAACAGC |
| F-CaDHCR7 | ATCTAAGTTTTAATTACAAAATGACCACCGCTGATGCT |
| R-CaDHCR7 | ATAAATCATAAGAAATTCGCTTAAAAAATATTTGGCAACAATCTATATGGAACAGC |
| F-XmDHCR7 | ATCTAAGTTTTAATTACAAAATGGAATCTACTAGAAGAAGACCTCAAC |
| R-XmDHCR7 | TTTTAAAACCTAAGAGTCACTGAACAGACCAGGAATCAATCTATGTG |
| F-CmDHCR7 | ATCTAAGTTTTAATTACAAAATGTCTAATCCATTTTTTGAATTACATCACCAG |
| R-CmDHCR7 | TTAATAATAAAAATCATAAAAAATAATCCAGGTAAAAGCCTCTGTGG |
| F-CgDHCR7 | ATCTAAGTTTTAATTACAAAATGATCGTATCAACTTGGTCCGG |
| R-CgDHCR7 | ACACTTATTTTTTTTATAACAAAGACACCAGGAATTAATCTGTCTGG |
| F-AmDHCR7 | ATCTAAGTTTTAATTACAAAATGTCTACAACCGAGAGTGTAAGG |
| R-AmDHCR7 | ACACTTATTTTTTTTATAACAAAAATCCCAGGTAACAATCTCTGC |
| F-TtDHCR7 | ATCTAAGTTTTAATTACAAAATGGCTTGTGATCAATATCAATGTTCT |
| R-TtDHCR7 | ATAAATCATAAGAAATTCGCTTAAAAAATACCTGGCAACAATCTATATGGAACAG |
| F-CcDHCR7 | ATCTAAGTTTTAATTACAAAATGACTACTGCAGATGCCGTC |
| R-CcDHCR7 | ATAAATCATAAGAAATTCGCGAAGATGTTAGGTAACAGTCTATAAGGCAC |
| F-ADH1t | TTTTTAAGCGAATTTCTTATGATTTATGATTTTTATTATTAAATAAGTTATAAAAAAAA |
| R- ADH1t | TGTCTGGACAAAGTTCTGTTTTTCCCCAGGAGTTAGCATATCTACAATTGGGTGAA |
| F-down-erg5 | CCCAATTGTAGATATGCTAACTCCTGGGGAAAAACAGAACTTTGTCCA |
| R-down-erg5 | TGACAGTGACGAACGCTTCAG |
| F-ACT1-qPCR | CTCCGTCTGGATTGGTGGTT |
| R-ACT1-qPCR | ACTTGTGGTGAACGATAGATGG |
| F-PhDHCR7-qPCR | TGTGATAGGCAGAGGCAAGAAT |
| R-PhDHCR7-qPCR | GGTGGTATAACTGGCGACGAT |
Table 2 Primers used in this study
| 引物 | 序列(5′→3′) |
|---|---|
| F-up-erg5 (delet) | TGGGAATACTGTACCAGATAATCAAACAT |
| R-up-erg5 (delet) | CAAAGTTCTGTTTTTCCCCATTTGTTAAAAGGTATTTATTGTCTATTGGAATAGC |
| F-down-erg5 (delet) | ATAAATACCTTTTAACAAATGGGGAAAAACAGAACTTTGTCCAGAC |
| R-down-erg5 (delet) | TGACAGTGACGAACGCTTCAG |
| F-erg5 | ATGAGTTCTGTCGCAGAAAATATAATAC |
| R-erg5 | TTATTCGAAGACTTCTCCAGTAATTGGG |
| F-up-erg5 | TGGGAATACTGTACCAGATAATCAAACATTAAA |
| R-up-erg5 | TGTTTATACGCTATTATCAGCCAATTTGTTAAAAGGTATTTATTGTCTATTGGAATAGCA |
| F-GAL1p | CCAATAGACAATAAATACCTTTTAACAAATTGGCTGATAATAGCGTATAAACAATGCA |
| R- GAL1p | TAACTCTATCAGAAGCCATCATTTTGTAATTAAAACTTAGATTAGATTGCTATGCTTTCT |
| F-DrDHCR7 | ATCTAAGTTTTAATTACAAAATGATGGCTTCTGATAGAGTTAGAAAAAG |
| R-DrDHCR7 | ATAAATCATAAGAAATTCGCTTAAAAAATATTTGGCAACAATCTATAAGAAACAGCAG |
| F-PhDHCR7 | ATCTAAGTTTTAATTACAAAATGTCTACCTCTGAAGGTGTTAGAAAAAG |
| R-PhDHCR7 | ATAAATCATAAGAAATTCGCTTAAAAAATACCTGGCAACAATCTATATCTAACGG |
| F-LrDHCR7 | ATCTAAGTTTTAATTACAAAATGGGTAGAGTTAAATGGAGATCTATTACC |
| R-LrDHCR7 | ATAAATCATAAGAAATTCGCTTAAAAAATATTTGGCAACAATCTATATGGAACAGC |
| F-CaDHCR7 | ATCTAAGTTTTAATTACAAAATGACCACCGCTGATGCT |
| R-CaDHCR7 | ATAAATCATAAGAAATTCGCTTAAAAAATATTTGGCAACAATCTATATGGAACAGC |
| F-XmDHCR7 | ATCTAAGTTTTAATTACAAAATGGAATCTACTAGAAGAAGACCTCAAC |
| R-XmDHCR7 | TTTTAAAACCTAAGAGTCACTGAACAGACCAGGAATCAATCTATGTG |
| F-CmDHCR7 | ATCTAAGTTTTAATTACAAAATGTCTAATCCATTTTTTGAATTACATCACCAG |
| R-CmDHCR7 | TTAATAATAAAAATCATAAAAAATAATCCAGGTAAAAGCCTCTGTGG |
| F-CgDHCR7 | ATCTAAGTTTTAATTACAAAATGATCGTATCAACTTGGTCCGG |
| R-CgDHCR7 | ACACTTATTTTTTTTATAACAAAGACACCAGGAATTAATCTGTCTGG |
| F-AmDHCR7 | ATCTAAGTTTTAATTACAAAATGTCTACAACCGAGAGTGTAAGG |
| R-AmDHCR7 | ACACTTATTTTTTTTATAACAAAAATCCCAGGTAACAATCTCTGC |
| F-TtDHCR7 | ATCTAAGTTTTAATTACAAAATGGCTTGTGATCAATATCAATGTTCT |
| R-TtDHCR7 | ATAAATCATAAGAAATTCGCTTAAAAAATACCTGGCAACAATCTATATGGAACAG |
| F-CcDHCR7 | ATCTAAGTTTTAATTACAAAATGACTACTGCAGATGCCGTC |
| R-CcDHCR7 | ATAAATCATAAGAAATTCGCGAAGATGTTAGGTAACAGTCTATAAGGCAC |
| F-ADH1t | TTTTTAAGCGAATTTCTTATGATTTATGATTTTTATTATTAAATAAGTTATAAAAAAAA |
| R- ADH1t | TGTCTGGACAAAGTTCTGTTTTTCCCCAGGAGTTAGCATATCTACAATTGGGTGAA |
| F-down-erg5 | CCCAATTGTAGATATGCTAACTCCTGGGGAAAAACAGAACTTTGTCCA |
| R-down-erg5 | TGACAGTGACGAACGCTTCAG |
| F-ACT1-qPCR | CTCCGTCTGGATTGGTGGTT |
| R-ACT1-qPCR | ACTTGTGGTGAACGATAGATGG |
| F-PhDHCR7-qPCR | TGTGATAGGCAGAGGCAAGAAT |
| R-PhDHCR7-qPCR | GGTGGTATAACTGGCGACGAT |
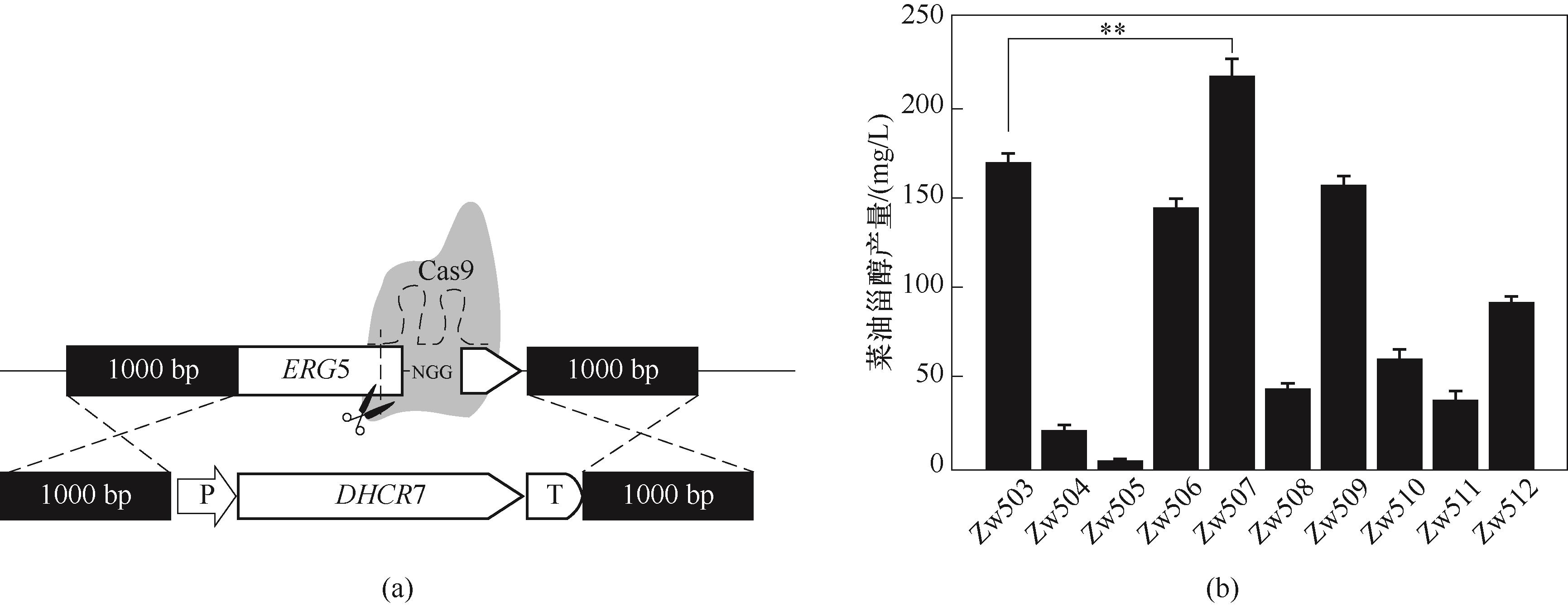
Fig.3 The construction of recombinant strains (a) and the production of DHCR7 campesterol from different species (b)( **p≤0.01 compared with the control strain; as determined by t test. The same below)
| 1 | Duport C, Spagnoli R, Degryse E, et al. Self-sufficient biosynthesis of pregnenolone and progesterone in engineered yeast[J]. Nature Biotechnology, 1998, 16 (2): 186-189. |
| 2 | Szczebara F M, Chandelier C, Villeret C, et al. Total biosynthesis of hydrocortisone from a simple carbon source in yeast[J]. Nature Biotechnology, 2003, 21 (2): 143-149. |
| 3 | Tsukagoshi Y, Suzuki H, Seki H, et al. Ajuga Δ24-sterol reductase catalyzes the direct reductive conversion of 24-methylenecholesterol to campesterol[J]. J. Biol. Chem., 2016, 291 (15): 8189-8198. |
| 4 | Zhao Y, Shen Y, Ma S, et al. Production of 5α-androstene-3,17-dione from phytosterols by co-expression of 5α-reductase and glucose-6-phosphate dehydrogenase in engineered Mycobacterium neoaurum[J]. Green Chemistry, 2019, 21 (7): 1809-1815. |
| 5 | Donova M V. Steroid bioconversions[J]. Methods Mol. Biol., 2017, 1645: 1-13. |
| 6 | Yao K, Xu L Q,Wang F Q, et al. Characterization and engineering of 3-ketosteroid-Δ1-dehydrogenase and 3-ketosteroid-9α-hydroxylase in Mycobacterium neoaurum ATCC 25795 to produce 9α-hydroxy-4-androstene-3,17-dione through the catabolism of sterols[J]. Metab. Eng., 2014, 24: 181-191. |
| 7 | Donova M V, Egorova O V. Microbial steroid transformations: current state and prospects[J]. Appl. Microbiol. Biotechnol., 2012, 94 (6): 1423-1447. |
| 8 | 张丽青.微生物转化在甾体药物合成中的应用[J].医药工业,1985, (1): 37-41. |
| Zhang L Q. Application of microbial transformation in the synthesis of steroid drugs [J]. Chinese Journal of Pharmaceuticals, 1985, (1): 37-41. | |
| 9 | Chen J, Fan F, Qu G, et al. Identification of Absidia orchidis steroid 11β-hydroxylation system and its application in engineering Saccharomyces cerevisiae for one-step biotransformation to produce hydrocortisone[J]. Metab. Eng., 2020, 57: 31-42. |
| 10 | Zhang W, Shao M, Rao Z, et al. Bioconversion of 4-androstene-3,17-dione to androst-1,4-diene-3,17-dione by recombinant Bacillus subtilis expressing ksdd gene encoding 3-ketosteroid-Δ1-dehydrogenase from Mycobacterium neoaurum JC-12[J]. The Journal of Steroid Biochemistry and Molecular Biology, 2013, 135: 36-42. |
| 11 | 吴玉玲,邵明龙,周武林, 等. 重组大肠杆菌表达17β-羟基类固醇脱氢酶全细胞催化合成宝丹酮的研究[J]. 化工学报, 2020, 71(7): 3229-3237. |
| Wu Y L, Shao M L, Zhou W L, et al. Study on catalytic synthesis of boldenone by recombinant Escherichia coli expressing 17β-hydroxysteroid dehydrogenase[J]. CIESC Journal, 2020, 71(7): 3229-3237. | |
| 12 | Ferreira R, Teixeira P G, Gossing M, et al. Metabolic engineering of Saccharomyces cerevisiae for overproduction of triacylglycerols[J]. Metab. Eng. Commun., 2018, 6: 22-27. |
| 13 | DiCarlo J E, Norville J E, Mali P, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems[J]. Nucleic Acids Res., 2013, 41 (7): 4336-4343. |
| 14 | Jakočiūnas T, Bonde I, Herrgård M, et al. Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae[J]. Metab. Eng., 2015, 28: 213-222. |
| 15 | Zalatan J G, Lee M E, Almeida R, et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds[J]. Cell, 2015, 160 (1): 339-350. |
| 16 | Lecain E, Chenivesse X, Spagnoli R, et al. Cloning by metabolic interference in yeast and enzymatic characterization of Arabidopsis thaliana sterol Δ7-reductase[J]. J. Biol. Chem., 1996, 271 (18): 10866-10873. |
| 17 | Du H X, Xiao W H, Wang Y, et al. Engineering Yarrowia lipolytica for campesterol overproduction[J]. PLoS One, 2016, 11 (1): e0146773. |
| 18 | Zhang Y, Wang Y, Yao M, et al. Improved campesterol production in engineered Yarrowia lipolytica strains[J]. Biotechnol. Lett., 2017, 39 (7): 1033-1039. |
| 19 | Wong J, d'Espaux L, Dev I, et al. De novo synthesis of the sedative valerenic acid in Saccharomyces cerevisiae[J]. Metab. Eng., 2018, 47: 94-101. |
| 20 | Reider Apel A, d'Espaux L,Wehrs M, et al. A Cas9-based toolkit to program gene expression in Saccharomyces cerevisiae[J]. Nucleic Acids Res., 2017, 45 (1): 496-508. |
| 21 | Li X, Roberti R, Blobel G. Structure of an integral membrane sterol reductase from Methylomicrobium alcaliphilum[J]. Nature, 2015, 517 (7532): 104-107. |
| 22 | Witsch-Baumgartner M, Löffler J, Utermann G. Mutations in thehuman DHCR7 gene[J]. Hum. Mutat., 2001, 17 (3): 172-182. |
| 23 | Prabhu A V, Luu W, Li D, et al. DHCR7: a vital enzyme switch between cholesterol and vitamin D production[J]. Prog. Lipid Res., 2016, 64: 138-151. |
| 24 | Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments[J]. Nucleic Acids Res., 2019, 47 (W1): W256-W259. |
| 25 | Li T, Liu G S, Zhou W, et al. Metabolic engineering of Saccharomyces cerevisiae to overproduce squalene[J]. J. Agric. Food Chem., 2020, 68 (7): 2132-2138. |
| 26 | Liu G S,Li T, Zhou W, et al. The yeast peroxisome: a dynamic storage depot and subcellular factory for squalene overproduction[J]. Metab. Eng., 2020, 57: 151-161. |
| 27 | Hubmann G, Thevelein J M, Nevoigt E. Natural and modified promoters for tailored metabolic engineering of the yeast Saccharomyces cerevisiae[J]. Methods Mol. Biol., 2014, 1152: 17-42. |
| 28 | Sun J, Shao Z, Zhao H, et al. Cloning and characterization of a panel of constitutive promoters for applications in pathway engineering in Saccharomyces cerevisiae[J]. Biotechnol. Bioeng., 2012, 109 (8): 2082-2092. |
| 29 | Monfort A, Finger S, Sanz P, et al. Evaluation of different promoters for the efficient production of heterologous proteins in baker's yeast[J]. Biotechnology Letters, 1999, 21 (3): 225-229. |
| 30 | Partow S, Siewers V, Bjørn S, et al. Characterization of different promoters for designing a new expression vector in Saccharomyces cerevisiae[J]. Yeast, 2010, 27 (11): 955-964. |
| 31 | Bhattacharya S, Esquivel B D, White T C. Overexpression or deletion of ergosterol biosynthesis genes alters doubling time, response to stress agents, and drug susceptibility in Saccharomyces cerevisiae[J]. mBio, 2018, 9 (4): e01291-18. |
| 32 | Liu J F, Xia J J, Nie K L, et al. Outline of the biosynthesis and regulation of ergosterol in yeast[J]. World J. Microbiol. Biotechnol., 2019, 35(7): 98. |
| 33 | Veen M, Stahl U, Lang C. Combined overexpression of genes of the ergosterol biosynthetic pathway leads to accumulation of sterols in Saccharomyces cerevisiae[J]. FEMS Yeast Research, 2003, 4 (1): 87-95. |
| 34 | Ma B X, Ke X, Tang X L, et al. Rate-limiting steps in the Saccharomyces cerevisiae ergosterol pathway: towards improved ergosta-5,7-dien-3β-ol accumulation by metabolic engineering[J]. World J. Microbiol. Biotechnol., 2018, 34 (4): 55. |
| 35 | 张振颖, 何秀萍, 李巍巍, 等. 甾醇C-24甲基转移酶和甾醇C-8异构酶在酿酒酵母麦角甾醇生物合成中的调控作用[J]. 微生物学报, 2009, 49 (8): 1063-1068. |
| Zhang Z Y, He X P, Li W W, et al. Regulation role of sterol C-24 methyltransferase and sterol C-8 isomerase in the ergosterol biosynthesis of Saccharomyces cerevisiae[J]. Acta Microbiologica Sinica, 2009, 49 (8): 1063-1068. |
| [1] | Xin LIU, Jun GE, Chun LI. Light-driven microbial hybrid systems improve level of biomanufacturing [J]. CIESC Journal, 2023, 74(1): 330-341. |
| [2] | Xue LIU, Lijuan ZHANG, Guangrong ZHAO. Commensalistic Escherichia coli coculture for biosynthesis of daidzein [J]. CIESC Journal, 2022, 73(9): 4015-4024. |
| [3] | Jingnan WANG, Jian PANG, Lei QIN, Chao GUO, Bo LYU, Chun LI, Chao WANG. Breeding and modification strategies of butenyl-spinosyn high-yield strains [J]. CIESC Journal, 2022, 73(2): 566-576. |
| [4] | Yi SUN, Teng ZHANG, Bo LYU, Chun LI. Improvement for fine regulation of microbial cell factory by intracellular biosensors [J]. CIESC Journal, 2022, 73(2): 521-534. |
| [5] | Xinhui WANG, Ying WANG, Mingdong YAO, Wenhai XIAO. Research progress of vitamin A biosynthesis [J]. CIESC Journal, 2022, 73(10): 4311-4323. |
| [6] | MAO Jinzhu, XIAO Shuling, YANG Zhichun, WANG Xiaoyu, ZHANG Shi, CHEN Junhong, XIE Jisheng, CHEN Fude, HUANG Zinuo, FENG Tianyu, ZHANG Aihui, FANG Baishan. Application of synthetic biology in pesticides residues detection [J]. CIESC Journal, 2021, 72(5): 2413-2425. |
| [7] | WANG Xin, ZHAO Peng, LI Qingyang, TIAN Pingfang. Research advances in semiconductor synthetic biology [J]. CIESC Journal, 2021, 72(5): 2426-2435. |
| [8] | WANG Lian, WU Di, ZHOU Jingwen. Research progress of lignans biosynthesis and their microbial production [J]. CIESC Journal, 2021, 72(1): 320-333. |
| [9] | ZHAO Zhenyao, ZHANG Baocai, LI Feng, SONG Hao. Design and construction of exoelectrogens by synthetic biology [J]. CIESC Journal, 2021, 72(1): 468-482. |
| [10] | WANG Kaifeng, WANG Jinpeng, WEI Ping, JI Xiaojun. Metabolic engineering of Yarrowia lipolytica to produce fatty acids and their derivatives [J]. CIESC Journal, 2021, 72(1): 351-365. |
| [11] | Hutao GAO, Xiaolin SHEN, Xinxiao SUN, Jia WANG, Qipeng YUAN. Metabolic engineering strategies in biosynthesis of amino acids and their derivatives [J]. CIESC Journal, 2020, 71(9): 4058-4070. |
| [12] | Jing XU, Zixuan YOU, Junqi ZHANG, Zheng CHEN, Deguang WU, Feng LI, Hao SONG. Advances in engineering electroactive biofilms by synthetic biology approaches [J]. CIESC Journal, 2020, 71(9): 3950-3962. |
| [13] | Lei QIN, Jie YU, Xiaoyu NING, Wentao SUN, Chun LI. Synthetic biological system construction and green intelligent biological manufacturing [J]. CIESC Journal, 2020, 71(9): 3979-3994. |
| [14] | Yanqin XU, Xizhi YANG, Ruoshi LUO, Yuhong HUANG, Feng HUO, Dan WANG. Application of synthetic biology in manufacture of bio-based plastics [J]. CIESC Journal, 2020, 71(10): 4520-4531. |
| [15] | Nuonan LI, Chun LI. Applications of glycosyltransferases in synthesis of triterpenoid saponins [J]. CIESC Journal, 2019, 70(10): 3869-3879. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||