化工学报 ›› 2024, Vol. 75 ›› Issue (7): 2624-2632.DOI: 10.11949/0438-1157.20240260
那雪梅1( ), 王雨1, 姜尧竹1, 贾男1, 王颖1(
), 王雨1, 姜尧竹1, 贾男1, 王颖1( ), 李春1,2
), 李春1,2
收稿日期:2024-03-04
修回日期:2024-05-14
出版日期:2024-07-25
发布日期:2024-08-09
通讯作者:
王颖
作者简介:那雪梅(1999—),女,硕士研究生,naxuemei@bit.edu.cn
基金资助:
Xuemei NA1( ), Yu WANG1, Yaozhu JIANG1, Nan JIA1, Ying WANG1(
), Yu WANG1, Yaozhu JIANG1, Nan JIA1, Ying WANG1( ), Chun LI1,2
), Chun LI1,2
Received:2024-03-04
Revised:2024-05-14
Online:2024-07-25
Published:2024-08-09
Contact:
Ying WANG
摘要:
熊果酸(UA)是具有多种生理及药理活性的植物来源五环三萜类羧酸,尽管在酿酒酵母中已经实现了UA的从头合成,但仍存在产量低、副产物积累、营养供给冗余等问题,无法实现工业化生产。为了提高酿酒酵母合成熊果酸的能力,通过多种方法优化了UA合成途径中异源CYP450酶的表达。探究了位置效应对CYP450基因表达的影响,初步筛选到CYP450的基因组最佳整合位点(HO位点),通过单独增加CYP450的拷贝数或同时增加CYP450及CPR的拷贝数,确定了同时存在3个CYP450和CPR拷贝时CYP450酶具有最佳催化活性,进行培养基优化后UA摇瓶产量达到(266.54±6.50) mg/L,最终在5 L发酵罐进行放大培养UA产量达到(2825.58±36.26)mg/L。本研究可为在酿酒酵母中通过表达优化策略调控其他三萜类化合物的合成提供参考。
中图分类号:
那雪梅, 王雨, 姜尧竹, 贾男, 王颖, 李春. 异源CYP450酶的表达优化促进工程酿酒酵母合成熊果酸[J]. 化工学报, 2024, 75(7): 2624-2632.
Xuemei NA, Yu WANG, Yaozhu JIANG, Nan JIA, Ying WANG, Chun LI. Expression optimization of heterologous CYP450 enzyme promotes the synthesis of ursolic acid in engineering Saccharomyces cerevisiae[J]. CIESC Journal, 2024, 75(7): 2624-2632.
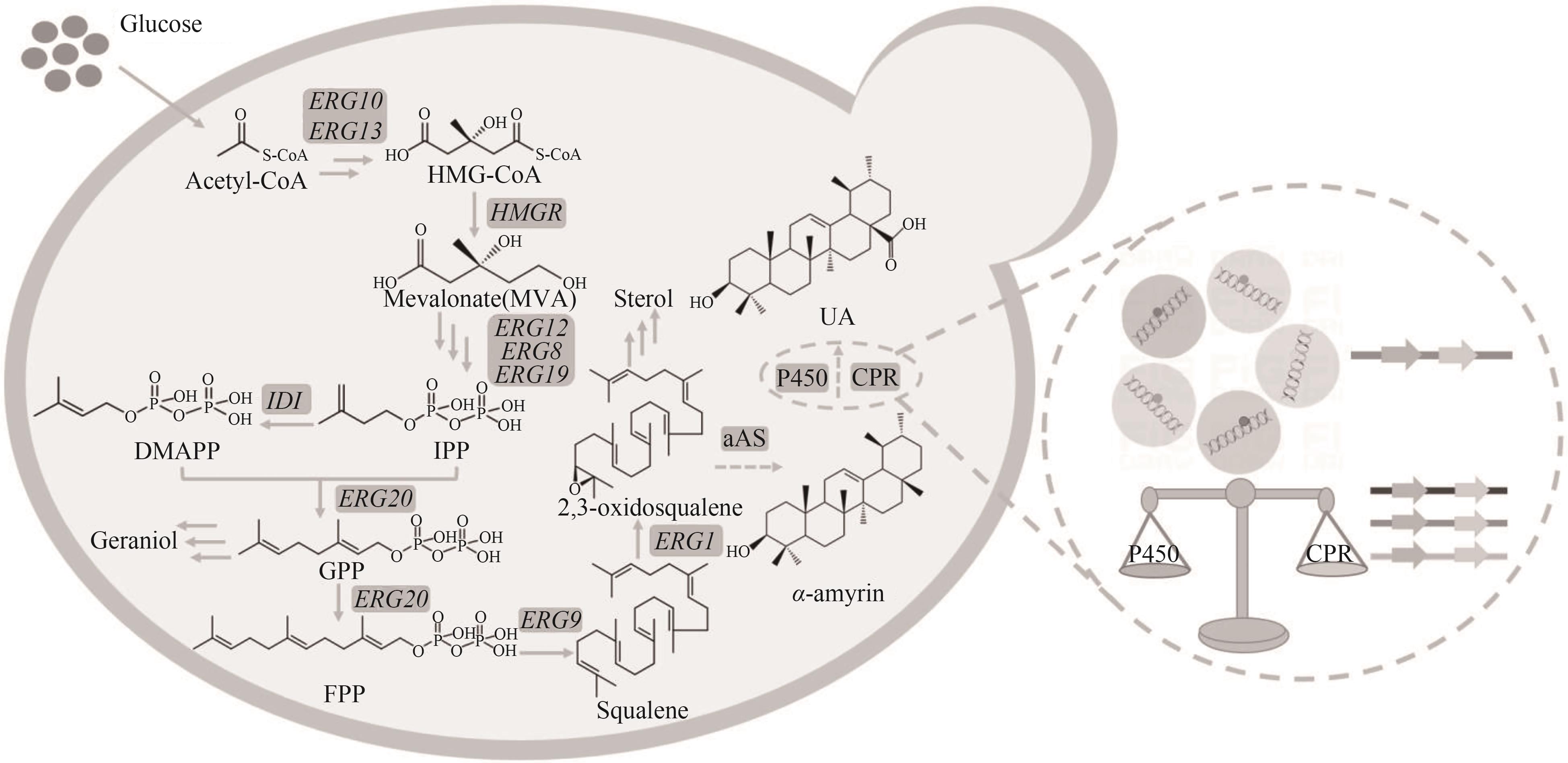
图1 酿酒酵母中熊果酸的合成途径及策略实线箭头代表内源途径,虚线箭头代表异源途径;Acetyl-CoA—乙酰辅酶A;ERG10—乙酰辅酶A转移酶基因;ERG13—HMG-CoA合酶基因;HMG-CoA—3-羟基-3-甲基戊二酰辅酶A;HMGR—HMG-CoA还原酶基因;ERG12—甲羟戊酸激酶基因;ERG8—磷酸甲羟戊酸激酶基因;ERG19—磷酸甲羟戊酸脱羧酶基因;IPP—异戊烯焦磷酸;IDI—IPP异构酶基因;DAMPP—二甲基烯丙基焦磷酸;ERG20—FPP合酶基因;GPP—香叶基焦磷酸;FPP—法尼基焦磷酸;ERG9—角鲨烯合酶基因;Squalene—角鲨烯;ERG1—角鲨烯环氧酶基因;2,3-oxidosqualene—2,3-氧化鲨烯;αAS—α-香树脂醇合酶;α-amyrin—α-香树脂醇;CYPC450—细胞色素P450酶;CPR—细胞色素还原酶;UA—熊果酸
Fig.1 Synthesis pathway and strategy of ursolic acid in S. cerevisiae
| Strain | Host strain | Genotype and [plasmid] | Source |
|---|---|---|---|
| DH5α | E. coli | F-φ80 lac ZΔM15 Δ(lacZYA-arg F) U169 endA1 recA1 hsdR17(rk-,mk+) supE44λ- thi -1 gyrA96 relA1 phoA | Tsingke |
| aAM12 | INVSc1 | MATa/MATα his3∆1 leu2 trp1-289 ura3-52Delta:: PGAL1-MdOSC1N11T/P250H_PGAL10-MdOSC1N11T/P250H_PADH1-DGA1-TDGA1_URA3r DNA::PTDH3-tHMG1_PALA1-ERG20_PGPM1 -ERG9_ PTYS1-ERG1_HIS3 [pESC-Trp-tHMG1-MdOSC1N11I/P250H] | this lab |
| XM-1 | aAM12 | HO::LEU2-CL8656-MtCPR | this lab |
| XM-2 | aAM12 | X-1::LEU2-CL8656-MtCPR | this lab |
| XM-3 | aAM12 | X-2::LEU2-CL8656-MtCPR | this lab |
| XM-4 | aAM12 | X-3::LEU2-CL8656-MtCPR | this lab |
| XM-5 | aAM12 | XI-2::LEU2-CL8656-MtCPR | this lab |
| XM-6 | aAM12 | XI-3::LEU2-CL8656-MtCPR | this lab |
| XM-7 | aAM12 | XI-4::LEU2-CL8656-MtCPR | this lab |
| XM-8 | aAM12 | XII-2::LEU2-CL8656-MtCPR | this lab |
| XM-9 | aAM12 | XII-3::LEU2-CL8656-MtCPR | this lab |
| XM-10 | aAM12 | XII-4::LEU2-CL8656-MtCPR | this lab |
| XM-11 | XM-1 | X-1::kanMX-CL8656 | this lab |
| XM-12 | XM-1 | rDNA::kanMX-CL8656-tHMG1-ERG20-ERG9-ERG1 | this lab |
| XM-13 | XM-1 | [pESC-kanMX-CL8656] | this lab |
| XM-14 | XM-1 | X-1::kanMX-CL8656-MtCPR | this lab |
| XM-15 | XM-14 | X-2:: BleoR-CL8656-MtCPR | this lab |
| XM-16 | XM-15 | [pRS415-Hygr-CL8656-MtCPR] | this lab |
| XM-17 | XM-15 | [pRS42K-Hygr-CL8656-MtCPR] | this lab |
表1 本研究中构建的菌株及质粒
Table 1 Strains and plasmids used in this study
| Strain | Host strain | Genotype and [plasmid] | Source |
|---|---|---|---|
| DH5α | E. coli | F-φ80 lac ZΔM15 Δ(lacZYA-arg F) U169 endA1 recA1 hsdR17(rk-,mk+) supE44λ- thi -1 gyrA96 relA1 phoA | Tsingke |
| aAM12 | INVSc1 | MATa/MATα his3∆1 leu2 trp1-289 ura3-52Delta:: PGAL1-MdOSC1N11T/P250H_PGAL10-MdOSC1N11T/P250H_PADH1-DGA1-TDGA1_URA3r DNA::PTDH3-tHMG1_PALA1-ERG20_PGPM1 -ERG9_ PTYS1-ERG1_HIS3 [pESC-Trp-tHMG1-MdOSC1N11I/P250H] | this lab |
| XM-1 | aAM12 | HO::LEU2-CL8656-MtCPR | this lab |
| XM-2 | aAM12 | X-1::LEU2-CL8656-MtCPR | this lab |
| XM-3 | aAM12 | X-2::LEU2-CL8656-MtCPR | this lab |
| XM-4 | aAM12 | X-3::LEU2-CL8656-MtCPR | this lab |
| XM-5 | aAM12 | XI-2::LEU2-CL8656-MtCPR | this lab |
| XM-6 | aAM12 | XI-3::LEU2-CL8656-MtCPR | this lab |
| XM-7 | aAM12 | XI-4::LEU2-CL8656-MtCPR | this lab |
| XM-8 | aAM12 | XII-2::LEU2-CL8656-MtCPR | this lab |
| XM-9 | aAM12 | XII-3::LEU2-CL8656-MtCPR | this lab |
| XM-10 | aAM12 | XII-4::LEU2-CL8656-MtCPR | this lab |
| XM-11 | XM-1 | X-1::kanMX-CL8656 | this lab |
| XM-12 | XM-1 | rDNA::kanMX-CL8656-tHMG1-ERG20-ERG9-ERG1 | this lab |
| XM-13 | XM-1 | [pESC-kanMX-CL8656] | this lab |
| XM-14 | XM-1 | X-1::kanMX-CL8656-MtCPR | this lab |
| XM-15 | XM-14 | X-2:: BleoR-CL8656-MtCPR | this lab |
| XM-16 | XM-15 | [pRS415-Hygr-CL8656-MtCPR] | this lab |
| XM-17 | XM-15 | [pRS42K-Hygr-CL8656-MtCPR] | this lab |
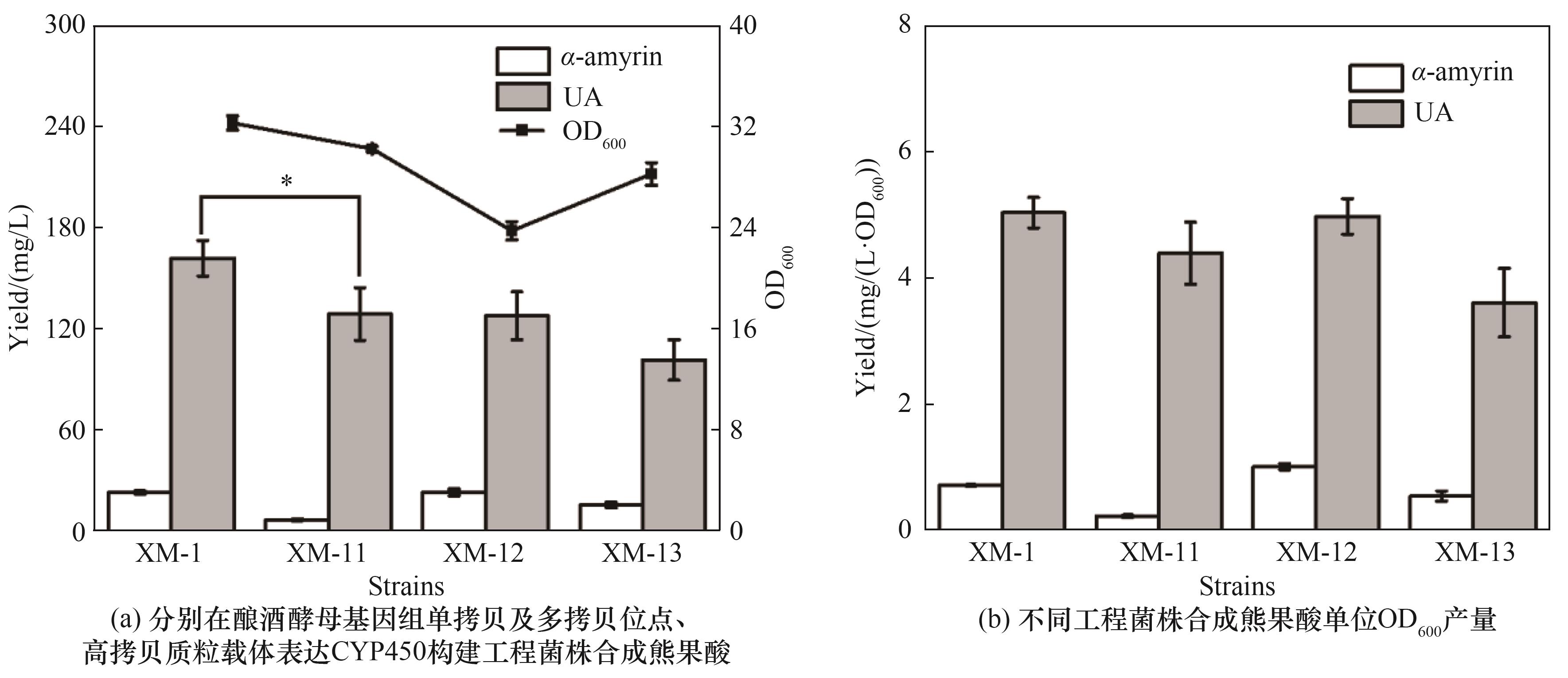
图3 单独增加CYP450拷贝数对酿酒酵母合成熊果酸的影响(误差线显示标准偏差并代表生物学重复,采用T检验*p<0.05,**p<0.01,***p<0.001进行统计分析)
Fig.3 Effect of increasing CYP450 copy number alone on ursolic acid synthesis by S. cerevisiae
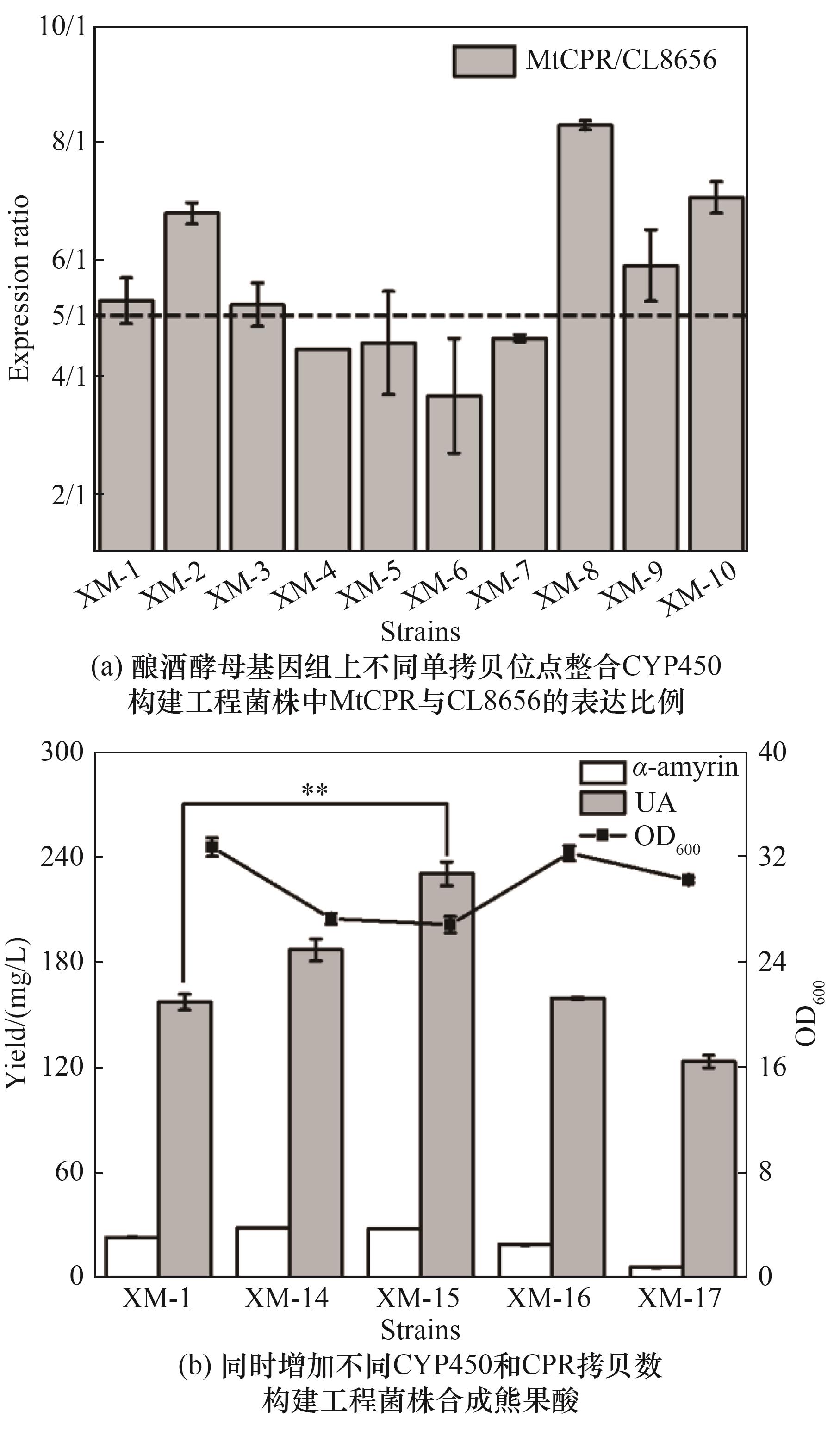
图4 同时增加CYP450和CPR拷贝数对酿酒酵母合成熊果酸的影响(误差线显示标准偏差并代表生物学重复,采用T检验*p<0.05,**p<0.01,***p<0.001进行统计分析)
Fig.4 Effect of simultaneous increase in CYP450 and CPR copy number on ursolic acid synthesis by S. cerevisiae
| C/N比 | 营养成分 | 金属离子 | α-amyrin产量/(mg/L) | UA产量/(mg/L) |
|---|---|---|---|---|
| 7∶1 | 2× | — | 34.63±0.72 | 236.99±3.37 |
| K+ | 40.04±3.78 | 266.54±6.50 | ||
| Fe2+ | 17.69±2.65 | 167.01±6.81 | ||
| 3× | — | 17.23±5.24 | 234.14±5.77 | |
| K+ | 23.87±2.39 | 266.21±7.27 | ||
| Fe2+ | 19.82±5.34 | 167.34±6.55 | ||
| 4× | — | 9.91±0.06 | 208.32±7.12 | |
| K+ | 14.97±0.62 | 239.00±8.57 | ||
| Fe2+ | 10.27±1.64 | 180.47±4.46 | ||
| 9∶1 | 2× | — | 23.13±0.96 | 237.56±8.82 |
| K+ | 33.48±0.80 | 225.51±5.74 | ||
| Fe2+ | 14.240±0.42 | 190.79±8.02 | ||
| 3× | — | 15.17±0.20 | 219.37±2.61 | |
| K+ | 17.77±0.20 | 163.11±2.56 | ||
| Fe2+ | 11.49±2.38 | 150.67±5.07 | ||
| 4× | — | 6.26±0.61 | 148.59±2.57 | |
| K+ | 8.32±0.21 | 144.50±5.99 | ||
| Fe2+ | 9.13±3.27 | 113.94±4.04 | ||
| 11∶1 | 2× | — | 23.58±1.35 | 168.71±5.05 |
| K+ | 35.01±2.76 | 163.66±2.26 | ||
| Fe2+ | 12.65±2.64 | 147.93±2.32 | ||
| 3× | — | 11.05±0.86 | 167.73±0.96 | |
| K+ | 12.48±1.01 | 142.91±4.78 | ||
| Fe2+ | 7.77±1.172 | 106.23±6.00 | ||
| 4× | — | 7.22±1.36 | 102.50±3.93 | |
| K+ | 6.74±0.053 | 99.96±5.74 | ||
| Fe2+ | 6.57±0.70 | 78.30±3.30 |
表2 XM-15菌株发酵条件优化
Table 2 Optimization of cultivation conditions for XM-15
| C/N比 | 营养成分 | 金属离子 | α-amyrin产量/(mg/L) | UA产量/(mg/L) |
|---|---|---|---|---|
| 7∶1 | 2× | — | 34.63±0.72 | 236.99±3.37 |
| K+ | 40.04±3.78 | 266.54±6.50 | ||
| Fe2+ | 17.69±2.65 | 167.01±6.81 | ||
| 3× | — | 17.23±5.24 | 234.14±5.77 | |
| K+ | 23.87±2.39 | 266.21±7.27 | ||
| Fe2+ | 19.82±5.34 | 167.34±6.55 | ||
| 4× | — | 9.91±0.06 | 208.32±7.12 | |
| K+ | 14.97±0.62 | 239.00±8.57 | ||
| Fe2+ | 10.27±1.64 | 180.47±4.46 | ||
| 9∶1 | 2× | — | 23.13±0.96 | 237.56±8.82 |
| K+ | 33.48±0.80 | 225.51±5.74 | ||
| Fe2+ | 14.240±0.42 | 190.79±8.02 | ||
| 3× | — | 15.17±0.20 | 219.37±2.61 | |
| K+ | 17.77±0.20 | 163.11±2.56 | ||
| Fe2+ | 11.49±2.38 | 150.67±5.07 | ||
| 4× | — | 6.26±0.61 | 148.59±2.57 | |
| K+ | 8.32±0.21 | 144.50±5.99 | ||
| Fe2+ | 9.13±3.27 | 113.94±4.04 | ||
| 11∶1 | 2× | — | 23.58±1.35 | 168.71±5.05 |
| K+ | 35.01±2.76 | 163.66±2.26 | ||
| Fe2+ | 12.65±2.64 | 147.93±2.32 | ||
| 3× | — | 11.05±0.86 | 167.73±0.96 | |
| K+ | 12.48±1.01 | 142.91±4.78 | ||
| Fe2+ | 7.77±1.172 | 106.23±6.00 | ||
| 4× | — | 7.22±1.36 | 102.50±3.93 | |
| K+ | 6.74±0.053 | 99.96±5.74 | ||
| Fe2+ | 6.57±0.70 | 78.30±3.30 |
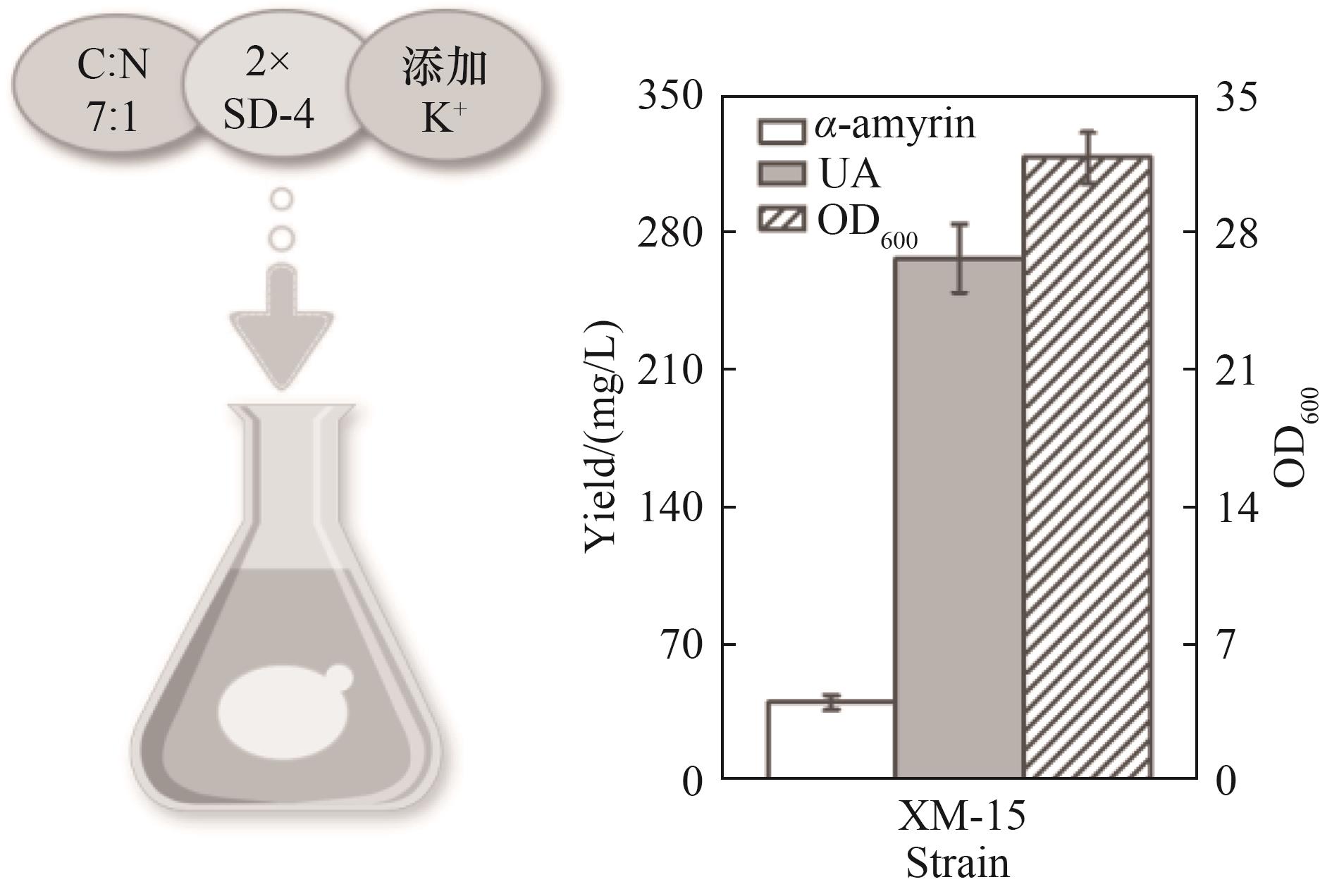
图5 XM-15菌株最优条件下摇瓶发酵合成熊果酸(所有数据均表示3个生物学重复的平均值,误差线显示标准偏差并代表生物学重复)
Fig.5 Ursolic acid was synthesized by shake flask fermentation of XM-15 strain under optimal conditions
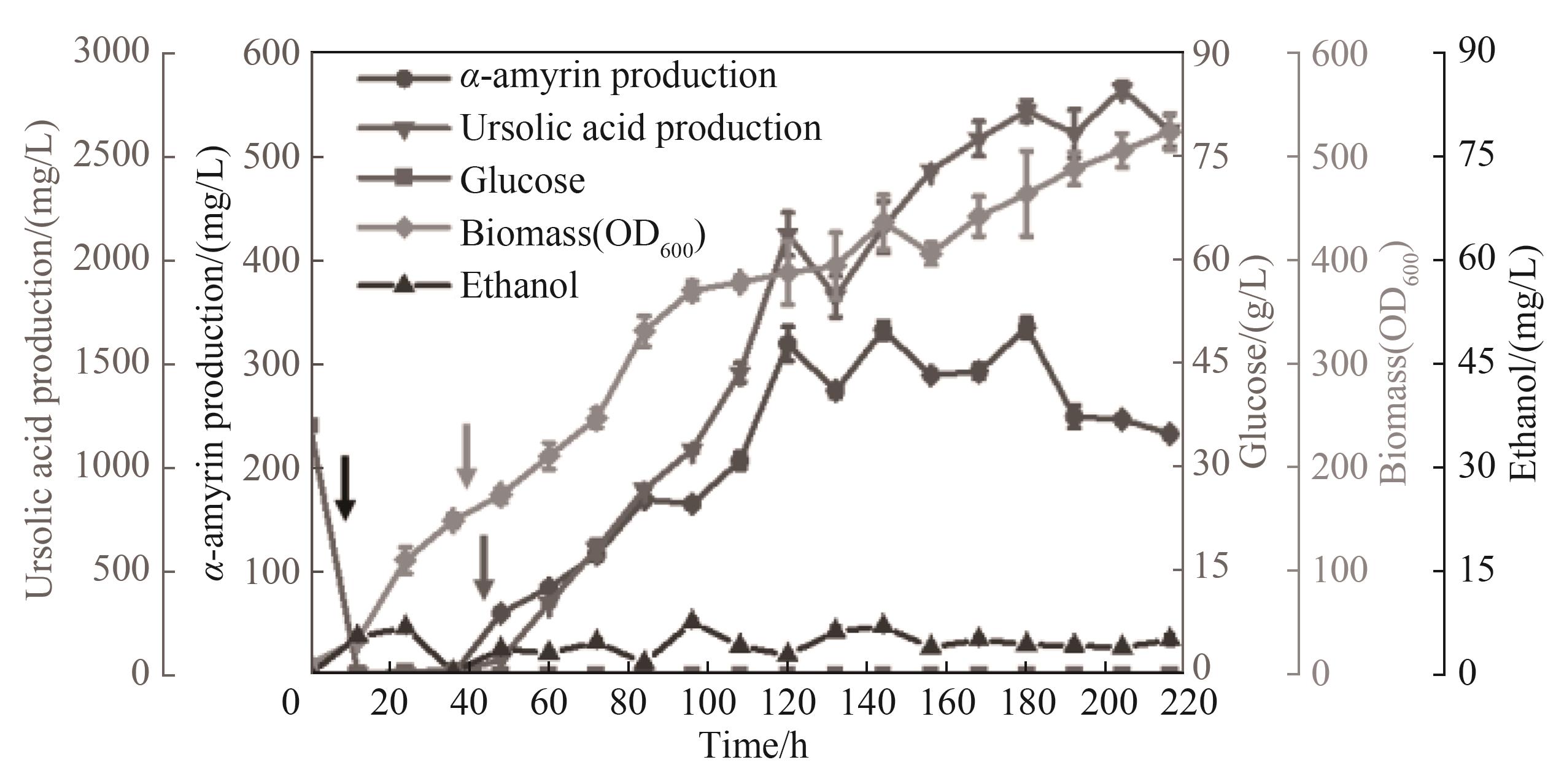
图6 XM-15菌株分批补料发酵合成熊果酸(所有数据均表示3个生物学重复的平均值,误差线显示标准偏差并代表生物学重复。从左往右第1个箭头表示葡萄糖开始补料,第2个箭头表示半乳糖开始补料,第3个箭头表示乙醇开始补料)
Fig.6 Ursolic acid was synthesized by fed-batch fermentation of XM-15 strain
| 1 | Zhu X X, Liu X N, Liu T, et al. Synthetic biology of plant natural products: from pathway elucidation to engineered biosynthesis in plant cells[J]. Plant Communications, 2021, 2(5): 100229. |
| 2 | Pemberton T A, Chen M B, Harris G G, et al. Exploring the influence of domain architecture on the catalytic function of diterpene synthases[J]. Biochemistry, 2017, 56(14): 2010-2023. |
| 3 | Lu C Z, Zhang C B, Zhao F L, et al. Biosynthesis of ursolic acid and oleanolic acid in Saccharomyces cerevisiae [J]. AIChE Journal, 2018, 64(11): 3794-3802. |
| 4 | Rashid S, Dar B A, Majeed R, et al. Synthesis and biological evaluation of ursolic acid-triazolyl derivatives as potential anti-cancer agents[J]. European Journal of Medicinal Chemistry, 2013, 66: 238-245. |
| 5 | Saravanan R, Viswanathan P, Pugalendi K V. Protective effect of ursolic acid on ethanol-mediated experimental liver damage in rats[J]. Life Sciences, 2006, 78(7): 713-718. |
| 6 | Yin R, Li T, Tian J X, et al. Ursolic acid, a potential anticancer compound for breast cancer therapy[J]. Critical Reviews in Food Science and Nutrition, 2018, 58(4): 568-574. |
| 7 | Balanehru S, Nagarajan B. Intervention of adriamycin induced free-radical damage[J]. Biochemistry International, 1992, 28(4): 735-744. |
| 8 | Xiang L P, Chi T, Tang Q, et al. A pentacyclic triterpene natural product, ursolic acid and its prodrug US597 inhibit targets within cell adhesion pathway and prevent cancer metastasis[J]. Oncotarget, 2015, 6(11): 9295-9312. |
| 9 | Charpe T W, Rathod V K. Separation of glycyrrhizic acid from licorice root extract using macroporous resin[J]. Food and Bioproducts Processing, 2015, 93: 51-57. |
| 10 | Cravens A, Payne J, Smolke C D. Synthetic biology strategies for microbial biosynthesis of plant natural products[J]. Nature Communications, 2019, 10(1): 2142. |
| 11 | Guo H, Wang H Y, Huo Y X. Engineering critical enzymes and pathways for improved triterpenoid biosynthesis in yeast[J]. ACS Synthetic Biology, 2020, 9(9): 2214-2227. |
| 12 | Ikeda Y, Murakami A, Ohigashi H. Ursolic acid: an anti- and pro-inflammatory triterpenoid[J]. Molecular Nutrition & Food Research, 2008, 52(1): 26-42. |
| 13 | Kong S J, Yu W, Gao N, et al. Expanding the neutral sites for integrated gene expression in Saccharomyces cerevisiae [J]. FEMS Microbiology Letters, 2022, 369(1): fnac081. |
| 14 | Velculescu V E, Zhang L, Zhou W, et al. Characterization of the yeast transcriptome[J]. Cell, 1997, 88(2): 243-251. |
| 15 | 张文政, 唐继军, 李炳志, 等. 酿酒酵母基因组位置效应对外源基因表达的影响[J]. 生物工程学报, 2016, 32(7): 901-911. |
| Zhang W Z, Tang J J, Li B Z, et al. Effect of integration loci of genome on heterologous gene expression in Saccharomyces cerevisiae [J]. Chinese Journal of Biotechnology, 2016, 32(7): 901-911. | |
| 16 | Yamane S, Yamaoka M, Yamamoto M, et al. Region specificity of chromosome Ⅲ on gene expression in the yeast Saccharomyces cerevisiae [J]. The Journal of General and Applied Microbiology, 1998, 44(4): 275-281. |
| 17 | Bai F D, Siewers V, Huang L, et al. Characterization of chromosomal integration sites for heterologous gene expression in Saccharomyces cerevisiae [J]. Yeast, 2009, 26(10): 545-551. |
| 18 | Thompson A, Gasson M J. Location effects of a reporter gene on expression levels and on native protein synthesis in Lactococcus lactis and Saccharomyces cerevisiae [J]. Applied and Environmental Microbiology, 2001, 67(8): 3434-3439. |
| 19 | Wu X L, Li B Z, Zhang W Z, et al. Genome-wide landscape of position effects on heterogeneous gene expression in Saccharomyces cerevisiae [J]. Biotechnology for Biofuels, 2017, 10(1): 189. |
| 20 | Urlacher V B, Girhard M. Cytochrome P450 monooxygenases in biotechnology and synthetic biology[J]. Trends in Biotechnology, 2019, 37(8): 882-897. |
| 21 | Cha Y P, Li W, Wu T, et al. Probing the synergistic ratio of P450/CPR to improve (+)-nootkatone production in Saccharomyces cerevisiae [J]. Journal of Agricultural and Food Chemistry, 2022, 70(3): 815-825. |
| 22 | Xu L P, Wang D, Chen J, et al. Metabolic engineering of Saccharomyces cerevisiae for gram-scale diosgenin production[J]. Metabolic Engineering, 2022, 70: 115-128. |
| 23 | Sun M C, Xin Q, Hou K X, et al. Production of 11-oxo-β-amyrin in Saccharomyces cerevisiae at high efficiency by fine-tuning the expression ratio of CYP450: CPR[J]. Journal of Agricultural and Food Chemistry, 2023, 71(8): 3766-3776. |
| 24 | 陈明凯, 叶丽丹, 于洪巍. 代谢改造酿酒酵母合成萜类化合物的研究进展[J]. 生物工程学报, 2021, 37(6): 2085-2104. |
| Chen M K, Ye L D, Yu H W. Advances in metabolic engineering of Saccharomyces cerevisiae for terpenoids biosynthesis[J]. Chinese Journal of Biotechnology, 2021, 37(6): 2085-2104. | |
| 25 | Wang S, Meng D, Feng M L, et al. Efficient plant triterpenoids synthesis in Saccharomyces cerevisiae: from mechanisms to engineering strategies[J]. ACS Synthetic Biology, 2024, 13(4): 1059-1076. |
| 26 | Carsanba E, Pintado M, Oliveira C. Fermentation strategies for production of pharmaceutical terpenoids in engineered yeast[J]. Pharmaceuticals, 2021, 14(4): 295. |
| 27 | Moses T, Pollier J, Almagro L, et al. Combinatorial biosynthesis of sapogenins and saponins in Saccharomyces cerevisiae using a C-16α hydroxylase from Bupleurum falcatum [J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(4): 1634-1639. |
| 28 | Jin K, Shi X, Liu J H, et al. Combinatorial metabolic engineering enables the efficient production of ursolic acid and oleanolic acid in Saccharomyces cerevisiae [J]. Bioresource Technology, 2023, 374: 128819. |
| 29 | Jia N, Li J Z, Zang G W, et al. Engineering Saccharomyces cerevisiae for high-efficient production of ursolic acid via cofactor engineering and acetyl-CoA optimization[J]. Biochemical Engineering Journal, 2024, 203: 109189. |
| 30 | 江丽红, 董昌, 黄磊, 等. 酿酒酵母代谢工程技术[J]. 生物工程学报, 2021, 37(5): 1578-1602. |
| Jiang L H, Dong C, Huang L, et al. Metabolic engineering tools for Saccharomyces cerevisiae [J]. Chinese Journal of Biotechnology, 2021, 37(5): 1578-1602. |
| [1] | 孙涛, 孙美莉, 陆然, 余一梓, 王凯峰, 纪晓俊. 合成生物学改造酵母驱动丁二酸绿色生物制造[J]. 化工学报, 2024, 75(4): 1382-1393. |
| [2] | 高学金, 姚玉卓, 韩华云, 齐咏生. 基于注意力动态卷积自编码器的发酵过程故障监测[J]. 化工学报, 2023, 74(6): 2503-2521. |
| [3] | 赵春雷, 郭亮, 高聪, 宋伟, 吴静, 刘佳, 刘立明, 陈修来. 代谢工程改造大肠杆菌生产软骨素[J]. 化工学报, 2023, 74(5): 2111-2122. |
| [4] | 王喆, 王建林, 李季, 周新杰, 随恩光. 基于WSDPC-RVR的多模态间歇过程软测量方法[J]. 化工学报, 2023, 74(11): 4656-4669. |
| [5] | 贺巍, 曹永娜, 尚宏儒, 李崯雪, 郭超, 于艳玲. 生物质发酵余热回收系统优化设计与性能分析[J]. 化工学报, 2023, 74(10): 4302-4310. |
| [6] | 毕浩然, 张洋, 王凯, 徐晨晨, 霍奕影, 陈必强, 谭天伟. 微生物制造绿色化学品研究进展[J]. 化工学报, 2023, 74(1): 1-13. |
| [7] | 刘昕, 戈钧, 李春. 光驱动微生物杂合系统提高生物制造水平[J]. 化工学报, 2023, 74(1): 330-341. |
| [8] | 刘雪, 张莉娟, 赵广荣. 大肠杆菌偏利共培养系统合成大豆苷元[J]. 化工学报, 2022, 73(9): 4015-4024. |
| [9] | 王悦琳, 晁伟, 蓝晓程, 莫志朋, 佟淑环, 王铁峰. 合成气生物发酵法制乙醇的研究进展[J]. 化工学报, 2022, 73(8): 3448-3460. |
| [10] | 童海航, 石德智, 刘嘉宇, 蔡桦伊, 罗丹, 陈飞. 金属纳米颗粒辅助木质纤维素暗发酵生物制氢的研究进展[J]. 化工学报, 2022, 73(4): 1417-1435. |
| [11] | 高学金, 何紫鹤, 高慧慧, 齐咏生. 基于联合典型变量矩阵的多阶段发酵过程质量相关故障监测[J]. 化工学报, 2022, 73(3): 1300-1314. |
| [12] | 孙怡, 张腾, 吕波, 李春. 胞内生物传感器提高微生物细胞工厂的精细调控[J]. 化工学报, 2022, 73(2): 521-534. |
| [13] | 王靖楠, 庞建, 秦磊, 郭超, 吕波, 李春, 王超. 丁烯基多杀菌素高产菌株的选育和改造策略[J]. 化工学报, 2022, 73(2): 566-576. |
| [14] | 王欣慧, 王颖, 姚明东, 肖文海. 维生素A生物合成的研究进展[J]. 化工学报, 2022, 73(10): 4311-4323. |
| [15] | 刘海波, 王楠, 刘洪周, 陈铁柱, 李建昌. 电压扰动对EAD代谢通量中微生物与关键酶活性的影响[J]. 化工学报, 2022, 73(10): 4603-4612. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备 11010102001995号
京公网安备 11010102001995号