化工学报 ›› 2024, Vol. 75 ›› Issue (7): 2633-2643.DOI: 10.11949/0438-1157.20240356
收稿日期:2024-04-01
修回日期:2024-05-05
出版日期:2024-07-25
发布日期:2024-08-09
通讯作者:
郑仁朝
作者简介:吴哲明(1988—),男,博士,副教授,wuzheming@zjut.edu.cn
基金资助:
Zheming WU( ), Biyun ZHANG, Renchao ZHENG(
), Biyun ZHANG, Renchao ZHENG( )
)
Received:2024-04-01
Revised:2024-05-05
Online:2024-07-25
Published:2024-08-09
Contact:
Renchao ZHENG
摘要:
布瓦西坦是新一代抗癫痫药物,具有广阔的市场前景。利用腈水解酶立体选择性催化3-氰基己腈合成关键手性中间体(R)-3-氰基己酸,进一步经加氢、环化、手性拆分等合成布瓦西坦路线,具有原子经济性高等优势,然而目前挖掘的腈水解酶存在立体选择性差等缺陷。以腈水解酶PgNIT为研究对象,通过对其催化口袋和亚基界面改造获得了立体选择性大幅提高的突变体F164W/I202R,产物光学纯度由野生型的86%提升至97%,E值由17提高至111。分子动力学模拟研究表明,F164位点的改造减小了(R)-3-氰基己腈深入催化口袋内部的空间位阻,而“A”界面I202位点的改造增加了亚基之间缔合的距离和催化口袋区域的原子动态协同性,从而显著提升了腈水解酶的立体选择性。
中图分类号:
吴哲明, 张碧云, 郑仁朝. 腈水解酶立体选择性改造及其合成布瓦西坦[J]. 化工学报, 2024, 75(7): 2633-2643.
Zheming WU, Biyun ZHANG, Renchao ZHENG. Engineering of nitrilase enantioselectivity for efficient synthesis of brivaracetam[J]. CIESC Journal, 2024, 75(7): 2633-2643.
| Primer | Sequence(5′ to 3′) |
|---|---|
| T133-F | ACCCCA |
| T133-R | ACGTTCGAAGTG |
| E136-F | ACCCCAACTCACTTC |
| E136-R | CCCCAAATCATACG |
| F164-F | CTGGCGTGT |
| F164-R | TTATGTTCGAA |
| P188-F | TTCTGCGATGTAT |
| P188-R | CAAAAGCGCTGCC |
| F196-F | TGGCGAAGGT |
| F196-R | TACGCTGCGC |
| E201-F | GCGCAGCGTATG |
| E201-R | GACGAATGTTGAT |
| I202-F | GCAGCGTATGGAA |
| I202-R | GGCCTGACGAATGTT |
| N203-F | CGTATGGAAATC |
| N203-R | CGTGCTGACGAAT |
| I204-F | ATGGAAATCAAC |
| I204-R | GTGCGTGCTGACG |
| R205-F | GAAATCAACATT |
| R205-R | CCAGTGCGTGCTG |
| Q206-F | AAATCAACATTCGT |
| Q206-R | TCCAGTGCGTG |
表1 上下游引物
Table 1 Primers used in this work
| Primer | Sequence(5′ to 3′) |
|---|---|
| T133-F | ACCCCA |
| T133-R | ACGTTCGAAGTG |
| E136-F | ACCCCAACTCACTTC |
| E136-R | CCCCAAATCATACG |
| F164-F | CTGGCGTGT |
| F164-R | TTATGTTCGAA |
| P188-F | TTCTGCGATGTAT |
| P188-R | CAAAAGCGCTGCC |
| F196-F | TGGCGAAGGT |
| F196-R | TACGCTGCGC |
| E201-F | GCGCAGCGTATG |
| E201-R | GACGAATGTTGAT |
| I202-F | GCAGCGTATGGAA |
| I202-R | GGCCTGACGAATGTT |
| N203-F | CGTATGGAAATC |
| N203-R | CGTGCTGACGAAT |
| I204-F | ATGGAAATCAAC |
| I204-R | GTGCGTGCTGACG |
| R205-F | GAAATCAACATT |
| R205-R | CCAGTGCGTGCTG |
| Q206-F | AAATCAACATTCGT |
| Q206-R | TCCAGTGCGTG |
| 突变体 | Km/(mmol/L) | Kcat/min-1 | (Kcat/Km)/ (L/(mmol·min)) |
|---|---|---|---|
| PgNIT | 1.38±0.19 | 21.37±1.80 | 15.48 |
| F164W | 0.44±0.06 | 12.91±0.79 | 29.34 |
| F164W/I202R | 1.68±0.42 | 6.99±1.22 | 4.16 |
表2 PgNIT及其突变体的动力学参数
Table 2 Kinetic parameters of PgNIT and variants towards 3-cyanocapronitrile
| 突变体 | Km/(mmol/L) | Kcat/min-1 | (Kcat/Km)/ (L/(mmol·min)) |
|---|---|---|---|
| PgNIT | 1.38±0.19 | 21.37±1.80 | 15.48 |
| F164W | 0.44±0.06 | 12.91±0.79 | 29.34 |
| F164W/I202R | 1.68±0.42 | 6.99±1.22 | 4.16 |
| 突变体 | E | 比酶活/(U/mg) | 半衰期t1/2/h |
|---|---|---|---|
| PgNIT | 18±4 | 2.48±0.06 | 63.3±0.61 |
| F164W | 30±2 | 3.47±0.11 | 55.5±1.71 |
| F164W/I202R | 112±5 | 1.29±0.06 | 39.7±2.22 |
表3 PgNIT及其突变体的立体选择性、催化活性及热稳定性
Table 3 Kinetic parameters of PgNIT and variants towards 3-cyanocapronitrile
| 突变体 | E | 比酶活/(U/mg) | 半衰期t1/2/h |
|---|---|---|---|
| PgNIT | 18±4 | 2.48±0.06 | 63.3±0.61 |
| F164W | 30±2 | 3.47±0.11 | 55.5±1.71 |
| F164W/I202R | 112±5 | 1.29±0.06 | 39.7±2.22 |
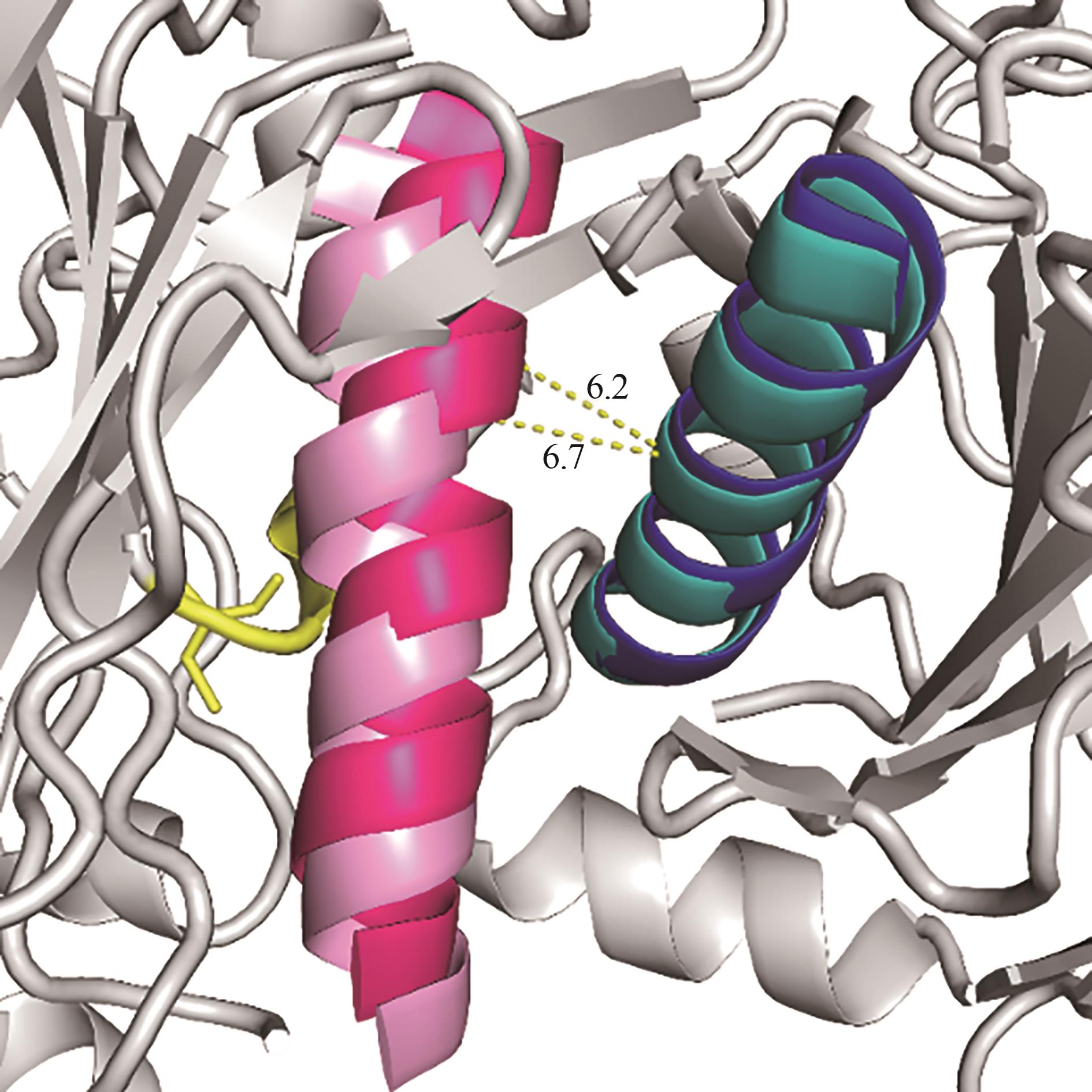
图10 PgNIT和F164W/I202R突变体的蛋白模型叠合图(黄色部分为C163,表示催化口袋位置;粉红色和浅蓝色Loop环为F164W/I202R的“A”界面关键Loop环;紫色和深蓝色Loop环为PgNIT的关键Loop环)
Fig.10 Illustration of protein model unity of PgNIT and its mutant F164W/I202R
| 1 | Rolan P, Sargentini-Maier M L, Pigeolet E, et al. The pharmacokinetics, cns pharmacodynamics and adverse event profile of brivaracetam after multiple increasing oral doses in healthy men[J]. British Journal of Clinical Pharmacology, 2008, 66(1): 71-75. |
| 2 | Markham A. Brivaracetam: first global approval[J]. Drugs, 2016, 76(4): 517-522. |
| 3 | Stephen L J, Brodie M J. Brivaracetam: a novel antiepileptic drug for focal-onset seizures[J]. Therapeutic Advances in Neurological Disorders, 2018, 11: 1-10. |
| 4 | 洪雪姣, 赵淑娟, 王漪檬, 等. 布瓦西坦的药理作用及临床评价[J]. 中国临床药理学杂志, 2017, 33(15): 1491-1493, 1502. |
| Hong X J, Zhao S J, Wang Y M, et al. Pharmacological effects and clinical evaluation of brivaracetam[J]. The Chinese Journal of Clinical Pharmacology, 2017, 33(15): 1491-1493, 1502. | |
| 5 | 杨君义, 李永法, 徐飞. 治疗部分性癫痫发作新药:布瓦西坦[J]. 中国新药与临床杂志, 2017, 36(1): 20-23. |
| Yang J Y, Li Y F, Xu F. New drug for partial epilepsies: brivaracetam[J]. Chinese Journal of New Drugs and Clinical Remedies, 2017, 36(1): 20-23. | |
| 6 | Russo E, Citraro R, Mula M. The preclinical discovery and development of brivaracetam for the treatment of focal epilepsy[J]. Expert Opinion on Drug Discovery, 2017, 12(11): 1169-1178. |
| 7 | Mercier J, Archen L, Bollu V, et al. Discovery of heterocyclic nonacetamide synaptic vesicle protein 2A (SV2A) ligands with single-digit nanomolar potency: opening avenues towards the first SV2A positron emission tomography (PET) ligands[J]. ChemMedChem, 2014, 9(4): 693-698. |
| 8 | Schülé A, Merschaert A, Szczepaniak C, et al. A biocatalytic route to the novel antiepileptic drug brivaracetam[J]. Organic Process Research & Development, 2016, 20(9): 1566-1575. |
| 9 | 郑仁朝, 郑裕国, 詹侃, 等. 一种酶法合成布瓦西坦手性中间体的方法: 114908075A[P]. 2022-08-16. |
| Zheng R C, Zheng Y G, Zhan K, et al. The invention relates to an enzymatic synthesis method of boisetan chiral intermediate: 114908075A [P]. 2022-08-16. | |
| 10 | Xue Y P, Shi C C, Xu Z, et al. Design of nitrilases with superior activity and enantioselectivity towards sterically hindered nitrile by protein engineering[J]. Advanced Synthesis & Catalysis, 2015, 357(8): 1741-1750. |
| 11 | Desantis G, Wong K, Farwell B, et al. Creation of a productive, highly enantioselective nitrilase through gene site saturation mutagenesis (GSSM)[J]. Journal of the American Chemical Society, 2003, 125(38): 11476-11477. |
| 12 | Yu S S, Li J L, Yao P Y, et al. Inverting the enantiopreference of nitrilase-catalyzed desymmetric hydrolysis of prochiral dinitriles by reshaping the binding pocket with a mirror-image strategy[J]. Angewandte Chemie International Edition, 2021, 60(7): 3679-3684. |
| 13 | Chen Z, Wang H L, Yang L, et al. Significantly enhancing the stereoselectivity of a regioselective nitrilase for the production of (S)-3-cyano-5-methylhexanoic acid using an MM/PBSA method[J]. Chemical Communications, 2021, 57(7): 931-934. |
| 14 | Gong J S, Lu Z M, Li H, et al. Nitrilases in nitrile biocatalysis: recent progress and forthcoming research[J]. Microbial Cell Factories, 2012, 11(1): 142. |
| 15 | Martínková L, Rucká L, Nešvera J, et al. Recent advances and challenges in the heterologous production of microbial nitrilases for biocatalytic applications[J]. World Journal of Microbiology and Biotechnology, 2017, 33(1): 8. |
| 16 | Banerjee A, Sharma R, Banerjee U C. A rapid and sensitive fluorometric assay method for the determination of nitrilase activity[J]. Biotechnology and Applied Biochemistry, 2003, 37(3): 289-293. |
| 17 | Lu X F, Diao H J, Wu Z M, et al. Engineering of reaction specificity, enantioselectivity, and catalytic activity of nitrilase for highly efficient synthesis of pregabalin precursor[J]. Biotechnology and Bioengineering, 2022, 119(9): 2399-2412. |
| 18 | Rakels J L, Straathof A J, Heijnen J J. A simple method to determine the enantiomeric ratio in enantioselective biocatalysis[J]. Enzyme and Microbial Technology, 1993, 15(12): 1051-1056. |
| 19 | Mirdita M, Schütze K, Moriwaki Y, et al. ColabFold: making protein folding accessible to all[J]. Nature Methods, 2022, 19(6): 679-682. |
| 20 | Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold[J]. Nature, 2021, 596(7873): 583-589. |
| 21 | Richard E, Michael O N, Alexander P, et al. Protein complex prediction with AlphaFold-Multimer[J]. Communications Biology, 2023, 6(1):1140. |
| 22 | Hooft R W W, Sander C, Vriend G. Objectively judging the quality of a protein structure from a ramachandran plot[J]. Bioinformatics, 1997, 13(4): 425-430. |
| 23 | Morris G M, Huey R, Lindstrom W, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility[J]. Journal of Computational Chemistry, 2009, 30(16): 2785-2791. |
| 24 | Berendsen H J C, van der Spoel D, van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation[J]. Computer Physics Communications, 1995, 91(1): 43-56. |
| 25 | Pronk S, Páll S, Schulz R, et al. Gromacs 4.5: a high-throughput and highly parallel open source molecular simulation toolkit[J]. Bioinformatics, 2013, 29(7): 845-854. |
| 26 | Lindorff-Larsen K, Piana S, Palmo K, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field[J]. Proteins, 2010, 78(8): 1950-1958. |
| 27 | Jorgensen W L, Chandrasekhar J, Madura J D, et al. Comparison of simple potential functions for simulating liquid water[J]. The Journal of Chemical Physics, 1983, 79(2): 926-935. |
| 28 | Liu Y X, Stone J E, Cai E, et al. VMD as a software for visualization and quantitative analysis of super resolution imaging and single particle tracking[J]. Biophysical Journal, 2014, 106(2): 202. |
| 29 | Seeliger D, de Groot B L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina[J]. Journal of Computer-Aided Molecular Design, 2010, 24(5): 417-422. |
| 30 | Li M, Liu X G, Zhang S L, et al. Deciphering the binding mechanism of inhibitors of the SARS-CoV-2 main protease through multiple replica accelerated molecular dynamics simulations and free energy landscapes[J]. Physical Chemistry Chemical Physics, 2022, 24(36): 22129-22143. |
| 31 | Thuku R N, Brady D, Benedik M J, et al. Microbial nitrilases: versatile, spiral forming, industrial enzymes[J]. Journal of Applied Microbiology, 2009, 106(3): 703-727. |
| 32 | Lu X F, Tang X L, Xu C H, et al. Engineering residues on C interface to improve thermostability of nitrilase for biosynthesis of pregabalin precursor[J]. AIChE Journal, 2023, 69(11): 18211. |
| 33 | Zhao L, Ma Z, Wang Q, et al. Engineering the thermostability of sucrose synthase by reshaping the subunit interaction contributes to efficient UDP-glucose production[J]. Journal of Agricultural and Food Chemistry, 2023, 71(8): 3832-3841. |
| 34 | Park J M, Mulelu A, Sewell B T, et al. Probing an interfacial surface in the cyanide dihydratase from bacillus pumilus, a spiral forming nitrilase[J]. Frontiers in Microbiology, 2016, 6: 1479. |
| 35 | Woodward J D, Trompetter I, Sewell B T, et al. Substrate specificity of plant nitrilase complexes is affected by their helical twist[J]. Communications Biology, 2018, 1(1): 186. |
| 36 | Yu H, Dalby P A. A beginner’s guide to molecular dynamics simulations and the identification of cross-correlation networks for enzyme engineering[J]. Methods in Enzymology, 2020, 643: 15-49. |
| 37 | Tang X L, Wen P F, Zheng W, et al. Bidirectional regulation of nitrilase reaction specificity by tuning the characteristic distances between key residues and substrate[J]. ACS Catalysis, 2023, 13(15): 10282-10294. |
| [1] | 秦晗淞, 李国梁, 闫昊, 冯翔, 刘熠斌, 陈小博, 杨朝合. 多级孔ZSM-5分子筛中油酸甲酯催化裂解吸附和扩散行为模拟研究[J]. 化工学报, 2024, 75(5): 1870-1881. |
| [2] | 刘东飞, 张帆, 刘铮, 卢滇楠. 机器学习势及其在分子模拟中的应用综述[J]. 化工学报, 2024, 75(4): 1241-1255. |
| [3] | 张政, 汪妩琼, 张雅静, 王康军, 吉远辉. 理论计算在药物制剂设计中的研究进展[J]. 化工学报, 2024, 75(4): 1429-1438. |
| [4] | 周康, 王建新, 于海, 魏朝良, 范丰奇, 车昕昊, 张磊. 基于分子动力学模拟的矿物基础油泡沫破裂性能研究[J]. 化工学报, 2024, 75(4): 1668-1678. |
| [5] | 李诗浩, 吴振华, 赵展烽, 吴洪, 杨冬, 石家福, 姜忠义. 化工过程中的电子传递、质子传递和分子传递[J]. 化工学报, 2024, 75(3): 1052-1064. |
| [6] | 吴凡, 彭旭东, 江锦波, 孟祥铠, 梁杨杨. 分子动力学模拟预测天然气密度和黏度的可行性研究[J]. 化工学报, 2024, 75(2): 450-462. |
| [7] | 宋明昊, 赵霏, 刘淑晴, 李国选, 杨声, 雷志刚. 离子液体脱除模拟油中挥发酚的多尺度模拟与研究[J]. 化工学报, 2023, 74(9): 3654-3664. |
| [8] | 胡建波, 刘洪超, 胡齐, 黄美英, 宋先雨, 赵双良. 有机笼跨细胞膜易位行为的分子动力学模拟研究[J]. 化工学报, 2023, 74(9): 3756-3765. |
| [9] | 赵佳佳, 田世祥, 李鹏, 谢洪高. SiO2-H2O纳米流体强化煤尘润湿性的微观机理研究[J]. 化工学报, 2023, 74(9): 3931-3945. |
| [10] | 程业品, 胡达清, 徐奕莎, 刘华彦, 卢晗锋, 崔国凯. 离子液体基低共熔溶剂在转化CO2中的应用[J]. 化工学报, 2023, 74(9): 3640-3653. |
| [11] | 汪林正, 陆俞冰, 张睿智, 罗永浩. 基于分子动力学模拟的VOCs热氧化特性分析[J]. 化工学报, 2023, 74(8): 3242-3255. |
| [12] | 陈吉, 洪泽, 雷昭, 凌强, 赵志刚, 彭陈辉, 崔平. 基于分子动力学的焦炭溶损反应及其机理研究[J]. 化工学报, 2023, 74(7): 2935-2946. |
| [13] | 陈雅鑫, 袁航, 刘冠章, 毛磊, 杨纯, 张瑞芳, 张光亚. 蛋白质纳米笼介导的酶自固定化研究进展[J]. 化工学报, 2023, 74(7): 2773-2782. |
| [14] | 汤晓玲, 王嘉瑞, 朱玄烨, 郑仁朝. 基于Pickering乳液的卤醇脱卤酶催化合成手性环氧氯丙烷[J]. 化工学报, 2023, 74(7): 2926-2934. |
| [15] | 董明, 徐进良, 刘广林. 超临界水非均质特性分子动力学研究[J]. 化工学报, 2023, 74(7): 2836-2847. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备 11010102001995号
京公网安备 11010102001995号