化工学报 ›› 2020, Vol. 71 ›› Issue (2): 724-735.DOI: 10.11949/0438-1157.20190862
收稿日期:2019-07-30
修回日期:2019-09-03
出版日期:2020-02-05
发布日期:2020-02-05
通讯作者:
卢滇楠
作者简介:彭雪(1994—),女,博士研究生,基金资助:
Xue PENG( ),Chenlin LU,Diannan LU(
),Chenlin LU,Diannan LU( )
)
Received:2019-07-30
Revised:2019-09-03
Online:2020-02-05
Published:2020-02-05
Contact:
Diannan LU
摘要:
为了揭示CO和O2竞争性结合人血红蛋白血红素位点的机制及其与人血红蛋白结构转换之间的关系,本文采用全原子分子动力学模拟(MD)结合马尔科夫状态模型(MSMs)研究氧气(O2)和一氧化碳(CO)分子从水溶液迁移进入人血红蛋白四聚体α链和β链的全过程。分子动力学模拟揭示了O2和CO结合α链和β链的稳态结合位点和瞬态结合位点、迁移通道以及α链的结构变化。结果显示,分子模拟不仅仅能够再现全部实验中所观察到的离散Xe结合位点和分子扩散通道,而且揭示了实验中无法观测的瞬态结合位点和多重气体迁移途径。上述结果表明人血红蛋白因其结构柔性所形成的瞬态通道对于气体分子迁移过程的重要性。除此之外,利用MSM和过渡路径理论(TPT)构建了人血红蛋白α链结构变化与气体分子迁移之间的关系,阐释了血红蛋白中影响气体迁移的关键结构及其微观机制。
中图分类号:
彭雪, 芦琛璘, 卢滇楠. 氧气和一氧化碳在人血红蛋白迁移过程研究[J]. 化工学报, 2020, 71(2): 724-735.
Xue PENG, Chenlin LU, Diannan LU. Investigation on migration process of oxygen and carbon monoxide in human hemoglobin[J]. CIESC Journal, 2020, 71(2): 724-735.
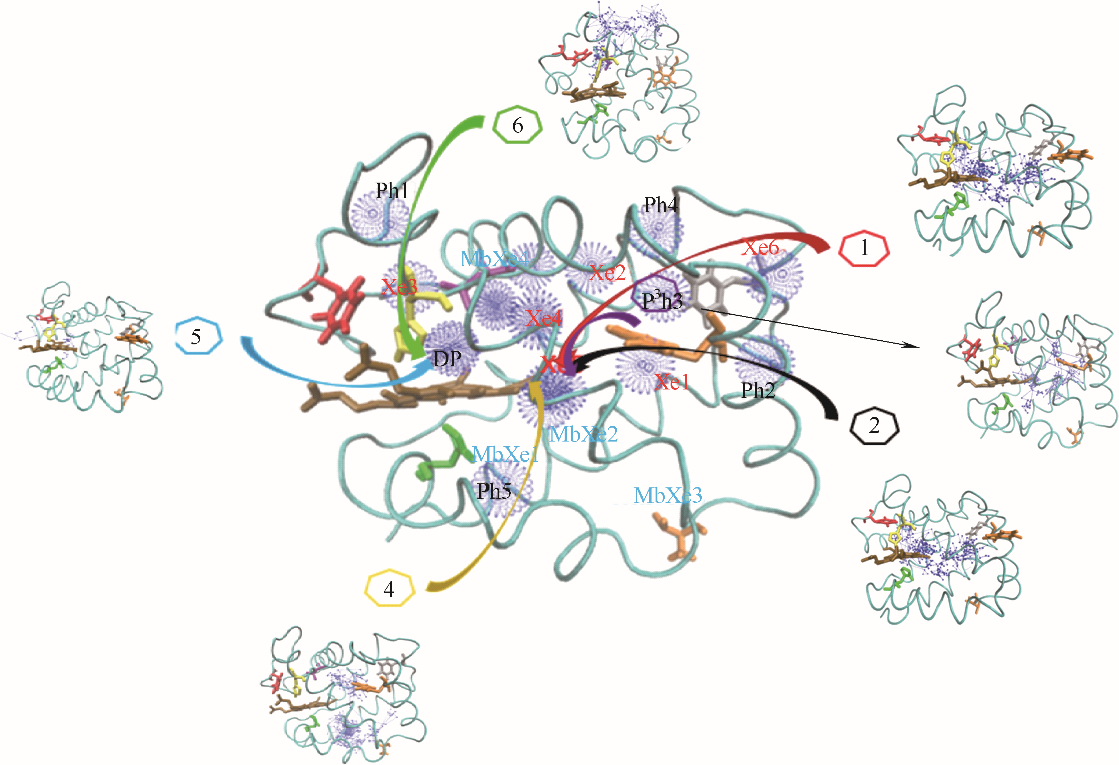
图1 O2在血红蛋白α链中的结合位点和六条主要迁移路径(蓝色球: O2位点;箭头连线: O2迁移轨迹;青色带:血红蛋白α链骨架;深棕色分子:血红素;Xe1~Xe6:实验确定Xe结合位点;DP:实验确定近端位点;Ph1~Ph5:瞬时空穴;MbXe1~MbXe4:实验确定肌红蛋白Xe结合位点)
Fig.1 O2 binding sites and 6 major migration pathways in α chain of human hemoglobin(blue ball: O2 sites; line with arrow: migration trajectories of O2; cyan belt: backbone of α chain of human hemoglobin; brown stick: heme; Xe1—Xe6: Xe binding sites determined by experiments; DP: distal path site determined by experiemnts; Ph1— Ph5: instant cavities; MbXe1 — MbXe4: Xe binding sites of myoglobin determined by experiments)
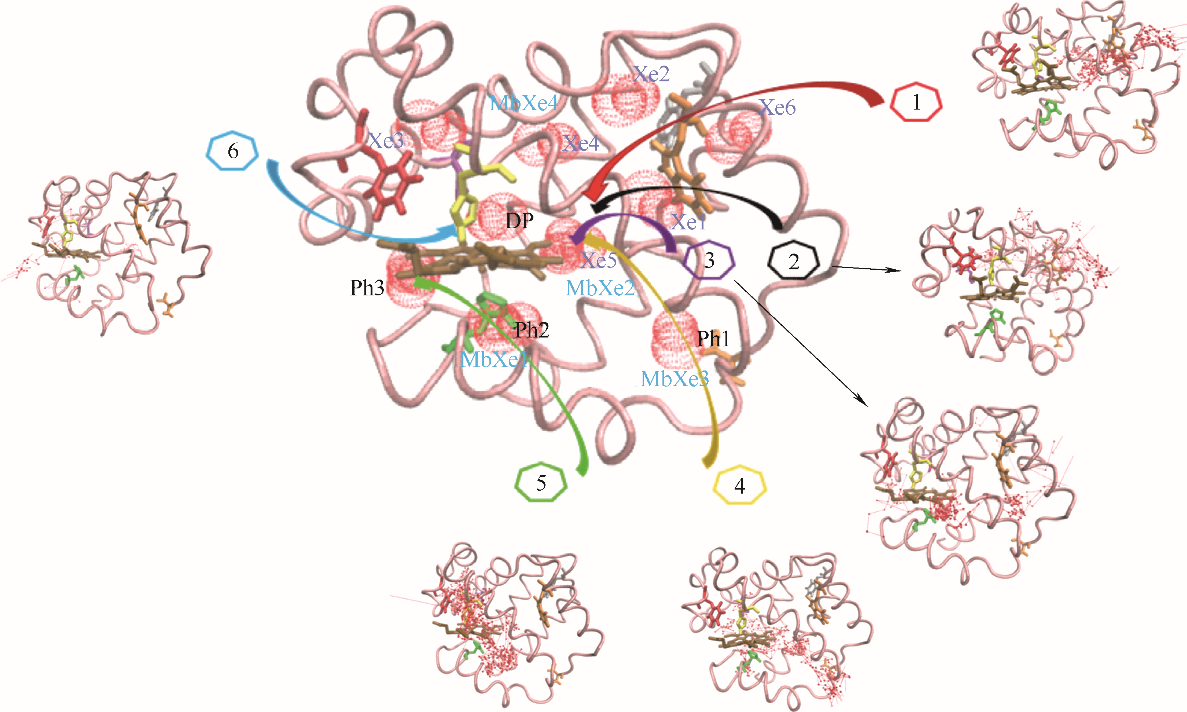
图4 CO在血红蛋白β链中的结合位点和六条主要迁移路径(红色球: CO位点;箭头连线: CO迁移轨迹;粉色带:血红蛋白β链骨架;深棕色分子:血红素;Xe1~Xe6:实验确定Xe结合位点;DP:实验确定近端位点;Ph1~Ph3:瞬时空穴;MbXe1~MbXe4:实验确定肌红蛋白Xe结合位点)
Fig.4 CO binding sites and 6 major migration pathways in β chain of human hemoglobin(red ball: CO sites; line with arrow: migration trajectories of CO; pink belt: backbone of β chain of human hemoglobin; brown stick: heme; Xe1—Xe6: Xe binding sites determined by experiments; DP: distal path site determined by experiemnts; Ph1—Ph3: instant cavities; MbXe1—MbXe4: Xe binding sites of myoglobin determined by experiments)
| 路径 | 次数 | 概率/% |
|---|---|---|
| 1: 水溶液→Xe6→Ph3→Xe5→DP | 49 | 49.0 |
| 2: 水溶液→Ph2→Xe1→Xe5→DP | 18 | 18.0 |
| 3: 水溶液→Ph3→Xe1→Xe5→DP | 3 | 3.0 |
| 4: 水溶液→Ph5→Xe5→Xe4→DP | 12 | 12.0 |
| 5: 水溶液→DP | 6 | 6.0 |
| 6: 水溶液→Ph1→Xe3→DP | 10 | 10.0 |
| 其他 | 2 | 2.0 |
表1 O2在α链中迁移路径的次数及概率
Table 1 Frequency and probability of O2 migration pathways in α chain
| 路径 | 次数 | 概率/% |
|---|---|---|
| 1: 水溶液→Xe6→Ph3→Xe5→DP | 49 | 49.0 |
| 2: 水溶液→Ph2→Xe1→Xe5→DP | 18 | 18.0 |
| 3: 水溶液→Ph3→Xe1→Xe5→DP | 3 | 3.0 |
| 4: 水溶液→Ph5→Xe5→Xe4→DP | 12 | 12.0 |
| 5: 水溶液→DP | 6 | 6.0 |
| 6: 水溶液→Ph1→Xe3→DP | 10 | 10.0 |
| 其他 | 2 | 2.0 |
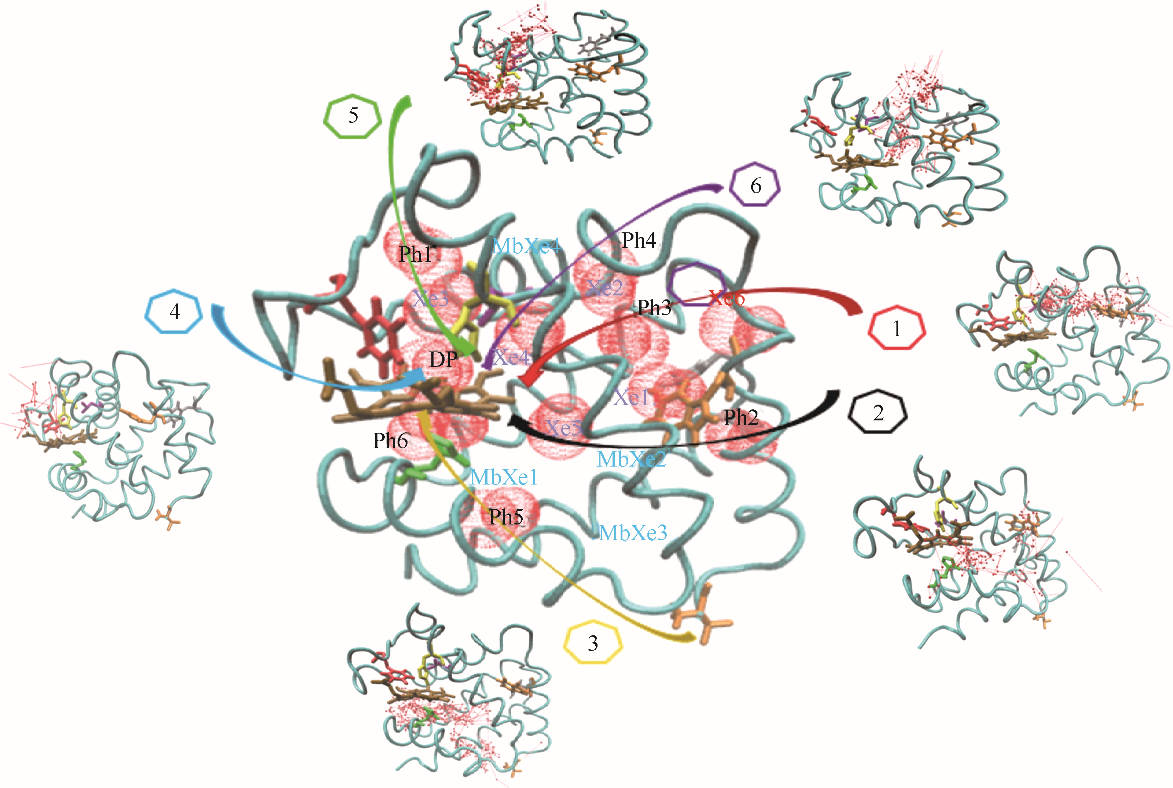
图2 CO在血红蛋白α链中的结合位点和六条主要迁移路径(红色球: CO位点;箭头连线: CO迁移轨迹;青色带:血红蛋白α链骨架;深棕色分子:血红素;Xe1~Xe6:实验确定Xe结合位点;DP:实验确定近端位点;Ph1~Ph6:瞬时空穴;MbXe1~MbXe4:实验确定肌红蛋白Xe结合位点)
Fig.2 CO binding sites and 6 major migration pathways in α chain of human hemoglobin(red ball: CO sites; line with arrow: migration trajectories of CO; cyan belt: backbone of α chain of human hemoglobin; brown stick: heme; Xe1—Xe6: Xe binding sites determined by experiments; DP: distal path site determined by experiemnts; Ph1—Ph6: instant cavities; MbXe1—MbXe4: Xe binding sites of myoglobin determined by experiment)
| 路径 | 次数 | 概率/% |
|---|---|---|
| 1: 水溶液→Xe6→Ph3→Xe4→DP | 21 | 17.9 |
| 2: 水溶液→Xe6→Ph2→Xe5→DP | 5 | 4.3 |
| 3: 水溶液→Ph5→Ph6→DP | 5 | 4.3 |
| 4: 水溶液→DP | 34 | 29.0 |
| 5: 水溶液→Ph1→Xe3→DP | 37 | 31.6 |
| 6: 水溶液→Ph4→Xe2→Xe4→DP | 12 | 10.3 |
| 其他 | 3 | 2.6 |
表2 CO在α链中迁移路径的次数及概率
Table 2 Frequency and probability of CO migration pathways in α chain
| 路径 | 次数 | 概率/% |
|---|---|---|
| 1: 水溶液→Xe6→Ph3→Xe4→DP | 21 | 17.9 |
| 2: 水溶液→Xe6→Ph2→Xe5→DP | 5 | 4.3 |
| 3: 水溶液→Ph5→Ph6→DP | 5 | 4.3 |
| 4: 水溶液→DP | 34 | 29.0 |
| 5: 水溶液→Ph1→Xe3→DP | 37 | 31.6 |
| 6: 水溶液→Ph4→Xe2→Xe4→DP | 12 | 10.3 |
| 其他 | 3 | 2.6 |

图3 O2在血红蛋白β链中的结合位点和六条主要迁移路径(蓝色球: O2位点;箭头连线: O2迁移轨迹;粉色带:血红蛋白β链骨架;深棕色分子:血红素;Xe1~Xe6:实验确定Xe结合位点;DP:实验确定近端位点;Ph1:瞬时空穴;MbXe1~MbXe4:实验确定肌红蛋白Xe结合位点)
Fig.3 O2 binding sites and 6 major migration pathways in β chain of human hemoglobin(blue ball: O2 sites; line with arrow: migration trajectories of O2; pink belt: backbone of β chain of human hemoglobin; brown stick: heme; Xe1—Xe6: Xe binding sites determined by experiments; DP: distal path site determined by experiemnts; Ph1: instant cavities; MbXe1 — MbXe4: Xe binding sites of myoglobin determined by experiments)
| 路径 | 次数 | 概率/% |
|---|---|---|
| 1: 水溶液→Xe6→Xe1→Xe5→DP | 25 | 25.0 |
| 2: 水溶液→Xe5→DP | 11 | 11.0 |
| 3: 水溶液→Xe1→Xe5→DP | 36 | 36.0 |
| 4: 水溶液→Ph1→Xe5→→DP | 5 | 5.0 |
| 5: 水溶液→DP | 4 | 4.0 |
| 6: 水溶液→Xe2→Xe4→DP | 16 | 16.0 |
| 其他 | 3 | 3.0 |
表3 O2在β链中迁移路径的次数及概率
Table 3 Frequency and probability of O2 migration pathways in β chain
| 路径 | 次数 | 概率/% |
|---|---|---|
| 1: 水溶液→Xe6→Xe1→Xe5→DP | 25 | 25.0 |
| 2: 水溶液→Xe5→DP | 11 | 11.0 |
| 3: 水溶液→Xe1→Xe5→DP | 36 | 36.0 |
| 4: 水溶液→Ph1→Xe5→→DP | 5 | 5.0 |
| 5: 水溶液→DP | 4 | 4.0 |
| 6: 水溶液→Xe2→Xe4→DP | 16 | 16.0 |
| 其他 | 3 | 3.0 |
| 路径 | 次数 | 概率/% |
|---|---|---|
| 1: 水溶液→Xe6→Xe1→Xe5→DP | 14 | 13.0 |
| 2: 水溶液→Xe1→Xe2→Xe4→DP | 18 | 16.7 |
| 3: 水溶液→Xe1→Xe5→DP | 7 | 6.5 |
| 4: 水溶液→Ph1→Xe5→DP | 18 | 16.7 |
| 5: 水溶液→Ph2→Ph3→DP | 7 | 6.4 |
| 6: 水溶液→DP | 40 | 37.0 |
| 其他 | 4 | 3.7 |
表4 CO在β链中迁移路径的次数及概率
Table 4 Frequency and probability of CO migration pathways in β chain
| 路径 | 次数 | 概率/% |
|---|---|---|
| 1: 水溶液→Xe6→Xe1→Xe5→DP | 14 | 13.0 |
| 2: 水溶液→Xe1→Xe2→Xe4→DP | 18 | 16.7 |
| 3: 水溶液→Xe1→Xe5→DP | 7 | 6.5 |
| 4: 水溶液→Ph1→Xe5→DP | 18 | 16.7 |
| 5: 水溶液→Ph2→Ph3→DP | 7 | 6.4 |
| 6: 水溶液→DP | 40 | 37.0 |
| 其他 | 4 | 3.7 |
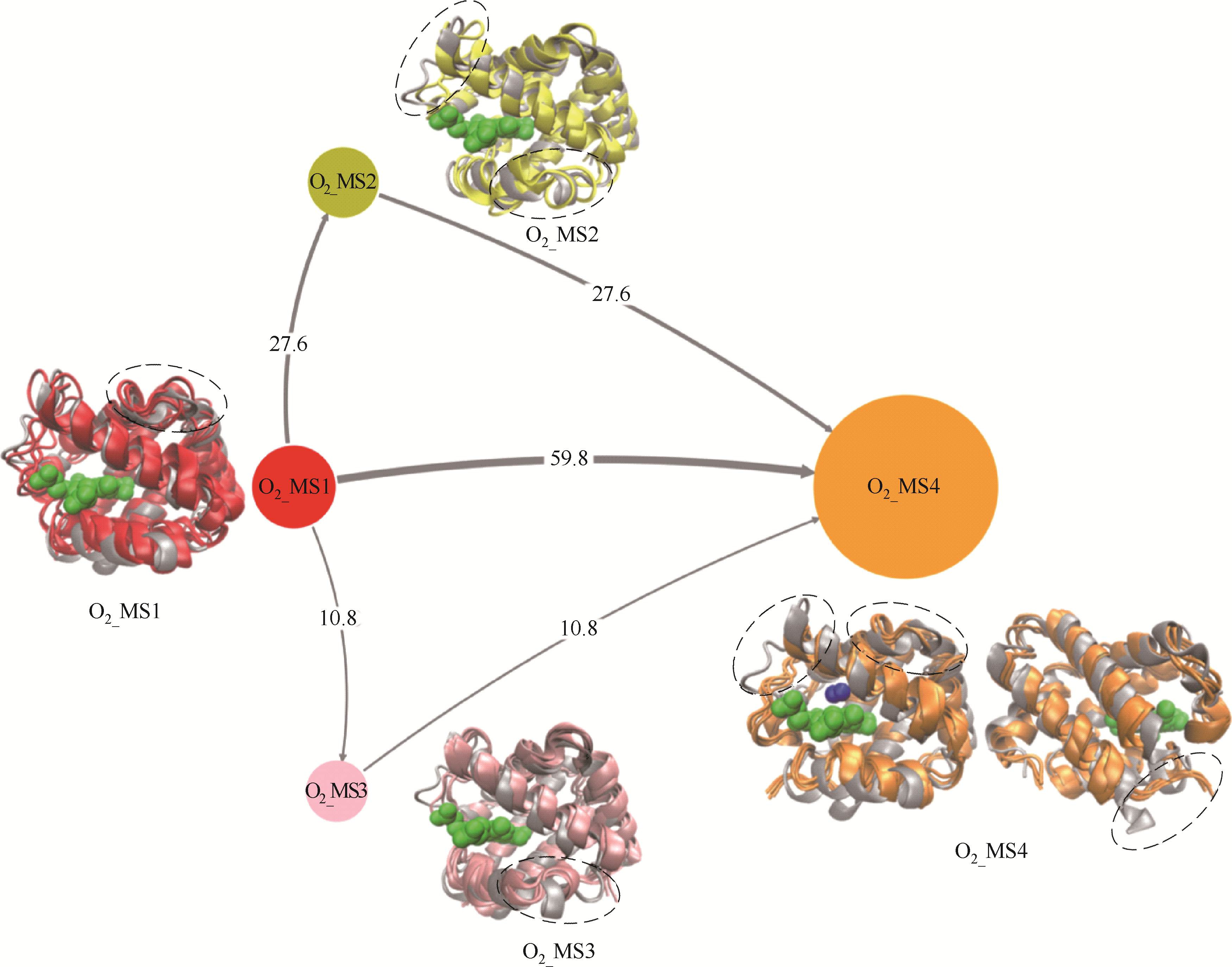
图7 O2迁移过程中人血红蛋白α链结构转换路径(灰色带状模型:人血红蛋白α链晶体结构;O2_MS1(红色): 初始状态结构,即O2未结合态; O2_MS2(黄色)和O2_MS3(粉色):中间状态结构,即迁移路径关键结构;O2_MS4(橙色):最终结合状态结构;含箭头连线:不同状态迁移路径,其粗细表明迁移概率;不同颜色圆:不同状态,其大小表明状态出现概率;不同颜色带状模型:对应状态人血红蛋白α链结构;绿色球状模型:血红素分子)
Fig.7 Tructural transition pathways of α chain of human hemoglobin during O2 migration processes(gray ribbon model: crystal structure of α chain of human hemoglobin; O2_MS1 in red: initial structure, i.e., O2 unbinding state; O2_MS2 in yellow and O2_MS3 in pink: middle states, i.e., key structures during O2 migration; O2_MS4 in orange: final O2 binding state; lines with arrow: transition pathways among different states, their thickness reflects transition probability; circles with different color: different states, their sizes reflect appearance probability; ribbon model with different color: corresponding structures of α chain of human hemoglobin; green ball model: heme molecule)
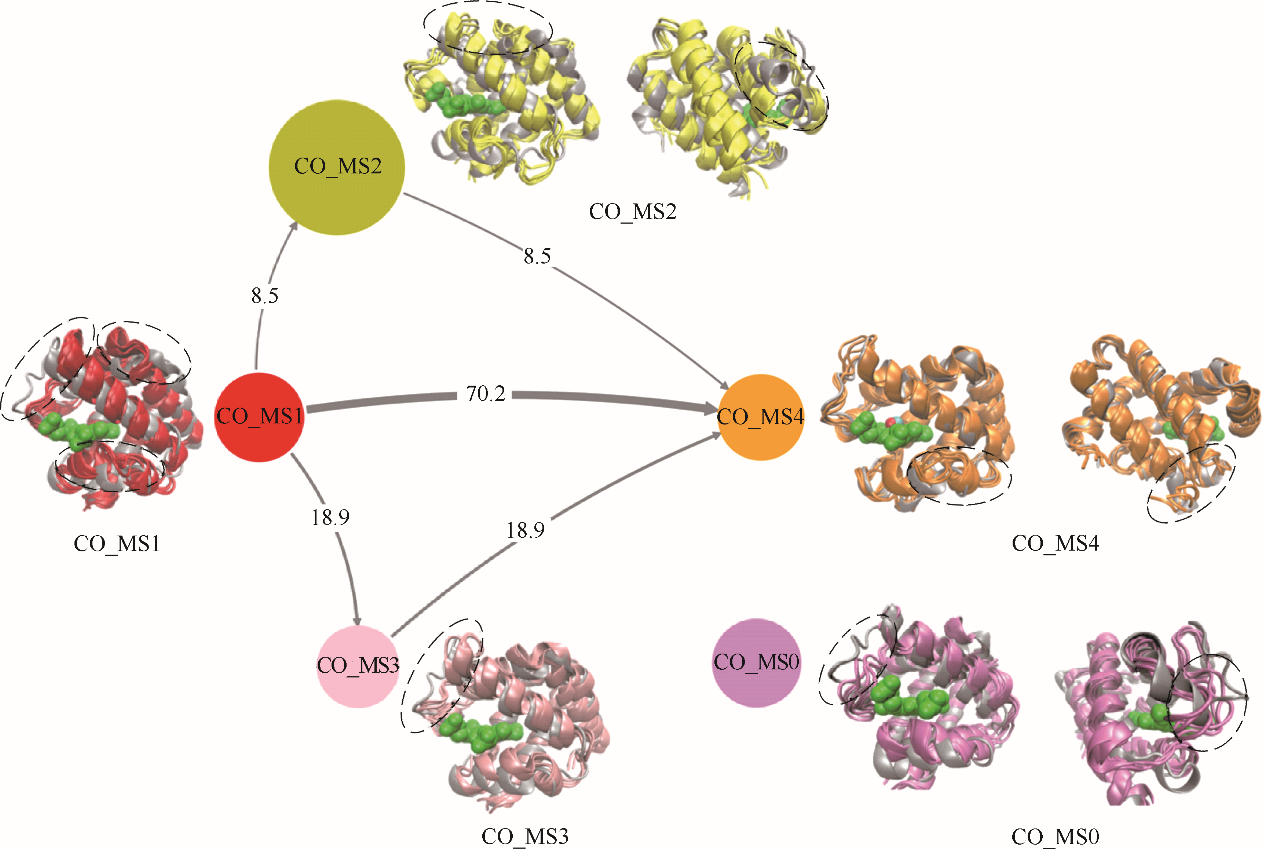
图8 CO迁移过程中人血红蛋白α链结构转换路径(灰色带状模型:人血红蛋白α链晶体结构;CO_MS1(红色): 初始状态结构,即CO未结合态;CO _MS2(黄色)和CO _MS3(粉色):中间状态结构,即迁移路径关键结构;CO _MS0(紫色):独立状态结构,即不参加转换的结构;CO _MS4(橙色):最终结合状态结构;含箭头连线:不同状态迁移路径,其粗细表明迁移概率;不同颜色圆:不同状态,其大小表明状态出现概率;不同颜色带状模型:对应状态人血红蛋白α链结构;绿色球状模型:血红素分子)
Fig.8 Structural transition pathways of α chain of human hemoglobin during CO migration processes(gray ribbon model: crystal structure of α chain of human hemoglobin; CO _MS1 in red: initial structure, i.e., CO unbinding state; CO _MS2 in yellow and CO _MS3 in pink: middle states, i.e., key structures during CO migration; CO _MS0 in purple: independent state, i.e., a structure not in transition network; CO _MS4 in orange: final CO binding state; lines with arrow: transition pathways among different states, their thickness reflects transition probability; circles with different color: different states, their sizes reflect appearance probability; ribbon model with different color: corresponding structures of α chain of human hemoglobin; green ball model: heme molecule)
| 1 | Feitelson J, McLendon G. Migration of small molecules through the structure of hemoglobin: evidence for gating in a protein electrontransfer reaction [J]. Bioc., 1991, 30: 5051-5055. |
| 2 | Leitner D M. Energy flow in proteins [J]. Annu. Rev. Phys. Chem., 2008, 59: 233-259. |
| 3 | Savino C, Miele A E, Draghi F, et al. Pattern of cavities in globins: the case of human hemoglobin [J]. Biopolymers, 2009, 91: 1097-1107. |
| 4 | Park S Y, Yokoyama T, Shibayama N, et al. 1.25 Å resolution crystal structures of human haemoglobin inthe oxy, deoxy and carbonmonoxy forms [J]. J. Mol. Biol., 2006, 360: 690-701. |
| 5 | Schotte F, Lim M, Jackson T, et al. Watching a protein as it functions with 150-ps time-resolved X-ray crystallography [J]. Science, 2003, 300: 1944-1947. |
| 6 | Schotte F, Soman J, Olson J S, et al. Picosecond time-resolved X-ray crystallography: probing protein function in real time [J]. Struct. Biol., 2004, 147: 235-246. |
| 7 | Schmidt M, Nienhaus K, Pahl R, et al. Ligand migration pathway and protein dynamics in myoglobin: a time-resolved crystallographic study on L29W MbCO [J]. Proc. Natl. Acad., 2005, 102: 11704-11709. |
| 8 | Perutz M F, Mathews F S. An X-ray study of azide methaemoglobin [J]. Biol., 1966, 21: 199- 202. |
| 9 | Lucas M F, Guallar V. An atomistic view on human hemoglobin carbon monoxide migration processes [J]. Biophys., 2012, 102: 887-896. |
| 10 | Lepeshkevich S V, Biziuk S A, Lemeza A M. The kinetics of molecular oxygen migration in the isolated alpha chains of human hemoglobin as revealed by molecular dynamics simulations and laser kinetic spectroscopy [J]. Biochimica et Biophysica Acta, 2011, 1814: 1279-1288. |
| 11 | Lepeshkevich S V, Gilevich S N, Parkhats M V, et al. Molecular oxygen migration through the xenon docking sites of human hemoglobin in the R-state [J]. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics, 2016, 1864(9): 1110-1121. |
| 12 | Hummer G, Schotte F, Anfinrud P A, et al. Unveiling functional protein motions with picosecond X-ray crystallography and molecular dynamics simulations [J]. Proceedings of the National Academy of Sciences of the United States of America, 2004, 101(43): 15330-15334. |
| 13 | Ruscio J Z, Kumar D, Shukla M. Atomic level computational identification of ligand migration pathways between solvent and binding site in myoglobin [J]. Proc. Natl., 2008, 105: 9204-9209. |
| 14 | Elber R. Ligand diffusion in globins simulations versu experiment [J]. Biol., 2010, 20: 162-167. |
| 15 | Ostermann A, Waschipky R, Parak F G, et al. Ligand binding and conformational motions in myoglobin [J]. Nature, 2000, 404(6774): 205-208. |
| 16 | Cordone L, Cottone G, Giuffrida S, et al. Thermal evolution of the CO stretching band in carboxy-myoglobin in the light of neutron scattering and molecular dynamics simulations [J]. Chemical Physics, 2008, 345(2): 275-282. |
| 17 | Yu T Q, Lapelosa M, Vandeneijnden E, et al. Full kinetics of CO entry, internal diffusion, and exit in myoglobin from transition-path theory simulations [J]. Journal of the American Chemical Society, 2015, 137(8): 3041-3050. |
| 18 | Alessandro B, Massimiliano B, Michele P. Wiley interdisciplinary reviews: computational molecular [J]. Journal of Computational Chemistry, 2011, 8: 826-843. |
| 19 | Leitner D M. Energy flow in proteins [J]. Annu. Rev. Phys. Chem., 2008, 59: 233-259. |
| 20 | Elber R, Karplus M. Enhanced sampling in molecular dynamics: use of the time-dependent hartree approximation for a simulation of carbon monoxide diffusion through myoglobin [J]. Journal of the American Chemical Society, 1990, 112: 9161-9175. |
| 21 | Meller J, Elber R. Computer simulations of carbon monoxide photodissociation in myoglobin: structural interpretation of the Bstates [J]. Biophys., 1998, 74: 789-802. |
| 22 | Bossa C, Anselmi M, Roccatano D, et al. Extended molecular dynamics simulation of thecarbon monoxide migration in sperm whale myoglobin [J]. Biophys., 2004, 86: 3855-3862. |
| 23 | Cohen J, Arkhipov A, Braun R, et al. Imaging the migration pathways for O2, CO, NO, and Xe inside myoglobin [J]. Biophysical Journal, 2006, 91(5): 1844-1857. |
| 24 | Sancho D D, Kubas A, Wang P H, et al. Identification of mutational hot spots for substrate diffusion: application to myoglobin [J]. Journal of Chemical Theory & Computation, 2015, 11(4): 1919-1927. |
| 25 | Shadrina M S, English A M, Peslherbe G H. Effective simulations of gas diffusion through kinetically accessible tunnels in multisubunit proteins: O2 pathways and escape routes in T-state deoxyhemoglobin [J]. J. Am. Chem. Soc., 2012, 134: 11177-11184. |
| 26 | Takayanagi M, Kurisaki I, Nagaoka M. Oxygen entry through multiple pathways in T-state human hemoglobin [J]. J. Phys. Chem. B, 2013, 117: 6082-6091. |
| 27 | Wang P H, Best R B, Blumberger J. Multiscale simulation reveals multiple pathways for H2 and O2 transport in a [NiFe]-hydrogenase [J]. Journal of the American Chemical Society, 2013, 10: 3548-3556. |
| 28 | Bustamante J P, Szretter M E, Sued M, et al. A quantitative model for oxygen uptake and release in a family of hemeproteins [J]. Bioinformatics, 2016, 32(12): 1805-1813. |
| 29 | Liang J, Edelsbrunner H, Fu P, et al. Analytical shape computation of macromolecules (Ⅱ): Inacessiblecavities in proteins [J]. Proteins: Struct. Func. Gen., 1998, 33: 18-29. |
| 30 | Shaanan B. Structure of human oxyhaemoglobin at 2.1 Å resolution [J]. Mol. Biol., 1983, 171: 31-59. |
| 31 | Wang P H, De S D, Best R B. Computation of rate constants for diffusion of small ligands to and from buried protein active sites [J]. Methods in Enzymology, 2016, 578: 299-326. |
| 32 | Birukou I, Maillett D H, Birukova A. Modulating distal cavities in the α and β subunits of human HbA reveals the primary ligand migration pathway [J]. Biochemistry, 2011, 50(34): 7361-7374. |
| 33 | Takayanagi M, Nagaoka M. Incipient structural and vibrational relaxation process of photolyzed carbonmonoxy myoglobin: statistical analysis by perturbation ensemble molecular dynamics method [J]. Theor. Chem. Acc., 2011, 130: 1115-1129. |
| 34 | Estarellas M C, Seira C C, Luque F J, et al. Understanding the kinetics of ligand binding to globins with molecular dynamics simulations: the necessity of multiple state models [J]. Drug Discovery Today Technologies, 2015, 17(70): 22-27. |
| 35 | Brooks B R, Bruccoleri R E, Olafson B D, et al. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations [J]. Comp. Chem., 1983, 4: 187-217. |
| 36 | Shadrina M S, Peslherbe G H, English A M. O2 and water migration pathways between the solvent and heme pockets of hemoglobin with open and closed conformations of the distal hisE7 [J]. Biochemistry, 2015, 54(34): 5279-5289. |
| 37 | Leitner D M, Straub J E. Proteins: Energy, Heat and Signal Flow (Computation in Chemistry) [M]. Boca Raton, FL: CRC Press, 2009. |
| 38 | Cameron A, Giovanni B. Enhanced sampling in molecular dynamics using metadynamics, replica exchange, and temperature-acceleration [J]. Entropy, 2013, 16: 163-199. |
| 39 | Berenbrink M. Evolution of vertebrate haemoglobins: histidine side chains, specific buffer value and Bohr effect [J]. Respiratory Physiology & Neurobiology, 2006, 154(1): 165-184. |
| 40 | Huber K P, Herzberg G. Molecular Spectra and Molecular Structure [M]. US: Springer, 1979. |
| [1] | 宋明昊, 赵霏, 刘淑晴, 李国选, 杨声, 雷志刚. 离子液体脱除模拟油中挥发酚的多尺度模拟与研究[J]. 化工学报, 2023, 74(9): 3654-3664. |
| [2] | 胡建波, 刘洪超, 胡齐, 黄美英, 宋先雨, 赵双良. 有机笼跨细胞膜易位行为的分子动力学模拟研究[J]. 化工学报, 2023, 74(9): 3756-3765. |
| [3] | 赵佳佳, 田世祥, 李鹏, 谢洪高. SiO2-H2O纳米流体强化煤尘润湿性的微观机理研究[J]. 化工学报, 2023, 74(9): 3931-3945. |
| [4] | 汪林正, 陆俞冰, 张睿智, 罗永浩. 基于分子动力学模拟的VOCs热氧化特性分析[J]. 化工学报, 2023, 74(8): 3242-3255. |
| [5] | 陈吉, 洪泽, 雷昭, 凌强, 赵志刚, 彭陈辉, 崔平. 基于分子动力学的焦炭溶损反应及其机理研究[J]. 化工学报, 2023, 74(7): 2935-2946. |
| [6] | 董明, 徐进良, 刘广林. 超临界水非均质特性分子动力学研究[J]. 化工学报, 2023, 74(7): 2836-2847. |
| [7] | 刘远超, 蒋旭浩, 邵钶, 徐一帆, 钟建斌, 李耑. 几何尺寸及缺陷对石墨炔纳米带热输运特性的影响[J]. 化工学报, 2023, 74(6): 2708-2716. |
| [8] | 顾浩, 张福建, 刘珍, 周文轩, 张鹏, 张忠强. 力电耦合作用下多孔石墨烯膜时间维度的脱盐性能及机理研究[J]. 化工学报, 2023, 74(5): 2067-2074. |
| [9] | 李辰鑫, 潘艳秋, 何流, 牛亚宾, 俞路. 基于碳微晶结构的炭膜模型及其气体分离模拟[J]. 化工学报, 2023, 74(5): 2057-2066. |
| [10] | 廖艺, 牛亚宾, 潘艳秋, 俞路. 复配表面活性剂对油水界面行为和性质影响的模拟研究[J]. 化工学报, 2022, 73(9): 4003-4014. |
| [11] | 杨松涛, 李东洋, 牛玉清, 李鑫钢, 康绍辉, 李洪, 叶开凯, 周志全, 高鑫. 氟化物势能函数和热力学性质的分子模拟研究进展[J]. 化工学报, 2022, 73(9): 3828-3840. |
| [12] | 陈晨, 杨倩, 陈云, 张睿, 刘冬. 不同氧浓度下煤挥发分燃烧的化学动力学研究[J]. 化工学报, 2022, 73(9): 4133-4146. |
| [13] | 刘振宇. 煤地下气化低效的化学反应工程根源:滞留层及通道中的传质与反应[J]. 化工学报, 2022, 73(8): 3299-3306. |
| [14] | 郑默, 李晓霞. ReaxFF MD模拟揭示的煤热解挥发分自由基反应的竞争与协调[J]. 化工学报, 2022, 73(6): 2732-2741. |
| [15] | 李春晖, 何辉, 何明键, 张萌, 高杨, 矫彩山. 离子液体萃取硝酸中Ce(Ⅳ)的动力学研究[J]. 化工学报, 2022, 73(4): 1606-1614. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备 11010102001995号
京公网安备 11010102001995号