化工学报 ›› 2022, Vol. 73 ›› Issue (9): 4015-4024.DOI: 10.11949/0438-1157.20220461
收稿日期:2022-03-30
修回日期:2022-05-30
出版日期:2022-09-05
发布日期:2022-10-09
通讯作者:
赵广荣
作者简介:刘雪(1992—),女,博士研究生,1208644284@qq.com基金资助:
Xue LIU1,2( ), Lijuan ZHANG1,2(
), Lijuan ZHANG1,2( ), Guangrong ZHAO1,2(
), Guangrong ZHAO1,2( )
)
Received:2022-03-30
Revised:2022-05-30
Online:2022-09-05
Published:2022-10-09
Contact:
Guangrong ZHAO
摘要:
大豆苷元是一种植物雌激素,具有多种生物活性,但在大肠杆菌中的生物全合成还未见报道。基于大豆苷元合成途径的三个模块(对香豆酸、甘草素和大豆苷元模块),构建大肠杆菌共培养系统从头合成大豆苷元。将对香豆酸和甘草素模块分配到两株大肠杆菌中构建双菌共培养系统,合成甘草素。在此基础上,探索了三种共培养模式合成大豆苷元,结果显示,三菌共培养系统比其他两种双菌共培养系统的产量更高,达到27.8 mg/L。共培养菌株间通过苯丙氨酸的单向流动形成了偏利共生的关系,有助于平衡代谢途径,提高大豆苷元产量。该共培养系统在大肠杆菌中实现大豆苷元的从头合成,为其他黄酮类化合物的生物合成提供了即插即用的平台。
中图分类号:
刘雪, 张莉娟, 赵广荣. 大肠杆菌偏利共培养系统合成大豆苷元[J]. 化工学报, 2022, 73(9): 4015-4024.
Xue LIU, Lijuan ZHANG, Guangrong ZHAO. Commensalistic Escherichia coli coculture for biosynthesis of daidzein[J]. CIESC Journal, 2022, 73(9): 4015-4024.
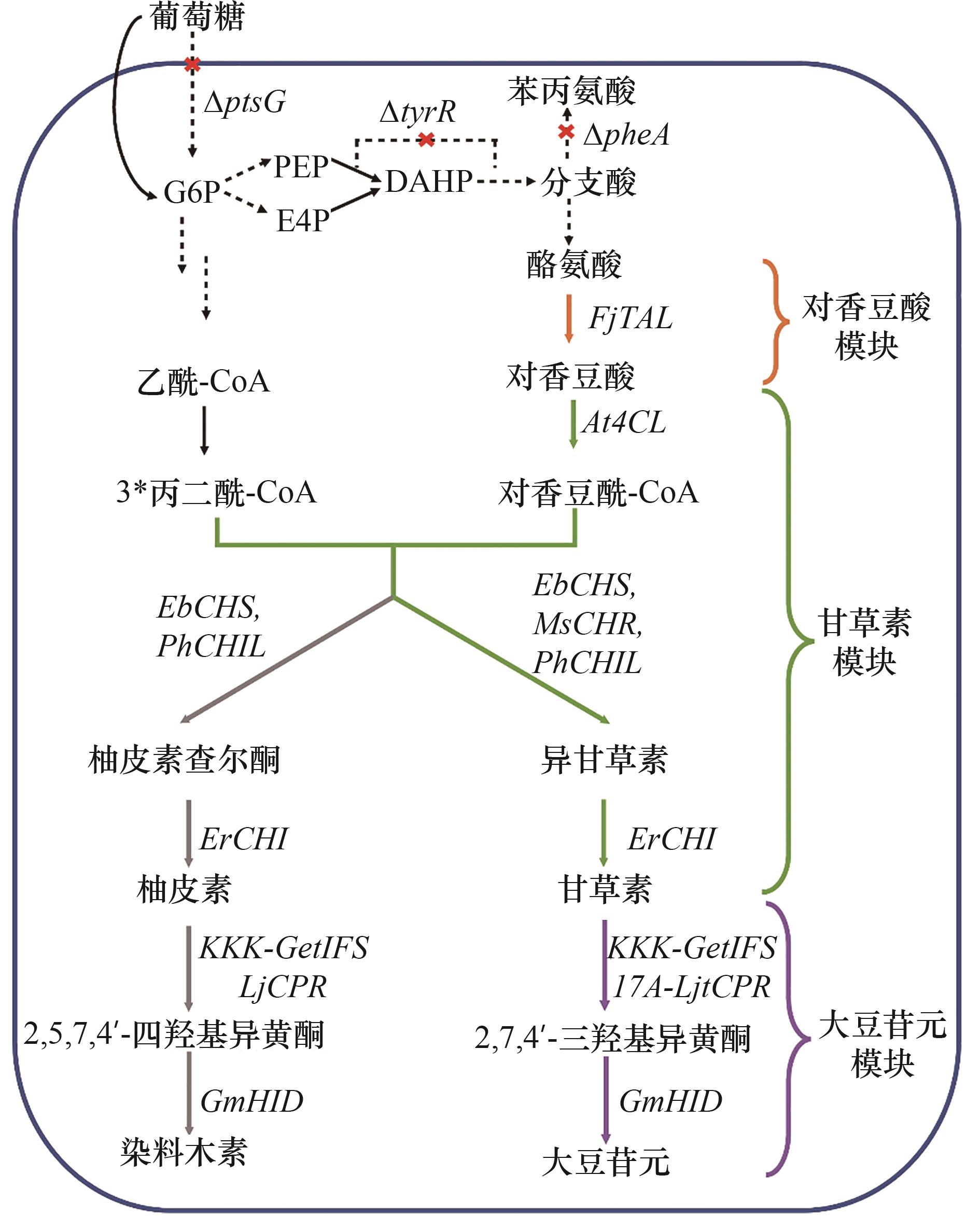
图1 代谢工程改造大肠杆菌从头合成大豆苷元红叉表示敲除的基因;实线箭头表示一步反应,虚线箭头表示多步反应,黑色箭头表示大肠杆菌内源途径,彩色箭头表示大豆苷元合成途径,灰色箭头表示副产物合成途径;G6P—葡萄糖6-磷酸;E4P—赤藓糖4-磷酸;PEP—磷酸烯醇式丙酮酸;DAHP—3-脱氧-D-阿拉伯庚酮糖酸-7-磷酸
Fig.1 Metabolic engineering E. coli for the de novo biosynthesis of daidzein
| Strains/plasmids | Characteristics | Source |
|---|---|---|
| strains | ||
| LTR3 | E. coli BL21(DE3) ΔptsG ΔtyrR ΔpheA::KanR | [ |
| LCA2G | LTR3 with pLCA1G and pYBT5C | [ |
| LLQ4 | BL21(DE3) with pLNR12 and pLLQ4 | this study |
| LLQ4R | BL21(DE3) with pLNR12R and pLLQ4 | this study |
| LLQ5 | LTR3 with pLNR13 and pLLQ4 | this study |
| LDZ8 | LTR3 with pLDZ1 and pLGN37 and pLLQ4 | this study |
| LDZ10 | BL21(DE3) with pLLQ4R, pLGN37K, and pLDZ4 | this study |
| LDZ11 | BL21(DE3) with pLGN37 and pLDZ3 | this study |
| LDZ12 | LTR3 with pLGN37 and pLDZ5 | this study |
| plasmids | ||
| pETDuet-1 | pET ori with PT7; AmpR | Novagen |
| pCDFDuet-1 | pCDF ori with PT7; SmR | Novagen |
| pACYCDuet-1 | pACYC ori with PT7; CmR | Novagen |
| pYBT5 | pMB1 ori with PlacUV5 -aroGfbr -tyrAfbr -aroE, Ptrc-ppsA-tktA-glk; AmpR | [ |
| pYBT5C | pYBT5 with change of AmpR to CmR | [ |
| pLCA1G | pCDF-FjTAL | [ |
| pLCA1G | pCDF-FjTAL-EGFP | [ |
| pLNR12 | pCDFDuet-1 with At4CL, PhCHIL | [ |
| pLNR12R | pCDFDuet-1 with At4CL, PhCHIL, and mCherry | [ |
| pLNR13 | pCDFDuet-1 with FjTAL, At4CL and PhCHIL | [ |
| pLLQ4 | pETDuet-1 with ErCHI and EbCHS-(GGGGGS)3-MsCHR | our lab |
| pLLQ4R | pETDuet-1 with ErCHI and EbCHS-(GGGGGS)3-MsCHR and mCherry | this study |
| pLGN37 | pACYCDuet-1 with 17A-LjtCPR | [ |
| pLGN37K | pLGN37 with exchange of CmR to KanR | [ |
| pLDZ1 | pCDFDuet-1 with PhCHIL, GmHID, FjTAL, At4CL and KKK-GetIFS | our lab |
| pLDZ3 | pCDFDuet-1 with KKK-GetIFS and GmHID | this study |
| pLDZ4 | pCDFDuet-1 with PhCHIL, GmHID, At4CL and KKK-GetIFS | our lab |
| pLDZ5 | pCDFDuet-1 with FjTAL, KKK-GetIFS, GmHID | this study |
表1 本研究所用菌株及质粒
Table 1 Strains and plasmids used in this study
| Strains/plasmids | Characteristics | Source |
|---|---|---|
| strains | ||
| LTR3 | E. coli BL21(DE3) ΔptsG ΔtyrR ΔpheA::KanR | [ |
| LCA2G | LTR3 with pLCA1G and pYBT5C | [ |
| LLQ4 | BL21(DE3) with pLNR12 and pLLQ4 | this study |
| LLQ4R | BL21(DE3) with pLNR12R and pLLQ4 | this study |
| LLQ5 | LTR3 with pLNR13 and pLLQ4 | this study |
| LDZ8 | LTR3 with pLDZ1 and pLGN37 and pLLQ4 | this study |
| LDZ10 | BL21(DE3) with pLLQ4R, pLGN37K, and pLDZ4 | this study |
| LDZ11 | BL21(DE3) with pLGN37 and pLDZ3 | this study |
| LDZ12 | LTR3 with pLGN37 and pLDZ5 | this study |
| plasmids | ||
| pETDuet-1 | pET ori with PT7; AmpR | Novagen |
| pCDFDuet-1 | pCDF ori with PT7; SmR | Novagen |
| pACYCDuet-1 | pACYC ori with PT7; CmR | Novagen |
| pYBT5 | pMB1 ori with PlacUV5 -aroGfbr -tyrAfbr -aroE, Ptrc-ppsA-tktA-glk; AmpR | [ |
| pYBT5C | pYBT5 with change of AmpR to CmR | [ |
| pLCA1G | pCDF-FjTAL | [ |
| pLCA1G | pCDF-FjTAL-EGFP | [ |
| pLNR12 | pCDFDuet-1 with At4CL, PhCHIL | [ |
| pLNR12R | pCDFDuet-1 with At4CL, PhCHIL, and mCherry | [ |
| pLNR13 | pCDFDuet-1 with FjTAL, At4CL and PhCHIL | [ |
| pLLQ4 | pETDuet-1 with ErCHI and EbCHS-(GGGGGS)3-MsCHR | our lab |
| pLLQ4R | pETDuet-1 with ErCHI and EbCHS-(GGGGGS)3-MsCHR and mCherry | this study |
| pLGN37 | pACYCDuet-1 with 17A-LjtCPR | [ |
| pLGN37K | pLGN37 with exchange of CmR to KanR | [ |
| pLDZ1 | pCDFDuet-1 with PhCHIL, GmHID, FjTAL, At4CL and KKK-GetIFS | our lab |
| pLDZ3 | pCDFDuet-1 with KKK-GetIFS and GmHID | this study |
| pLDZ4 | pCDFDuet-1 with PhCHIL, GmHID, At4CL and KKK-GetIFS | our lab |
| pLDZ5 | pCDFDuet-1 with FjTAL, KKK-GetIFS, GmHID | this study |
| Primers | Sequences(5'-3') |
|---|---|
| Cherry-CDF-F | GTCATCGTGGCCGGATCTTGCGCAAAAAACCCCTCAAGACC |
| Cherry-CDF-R | TTCGTTCAAGCCGAGGGGCCTCGAACAGAAAGTAATCGTA |
| CDF-Cherry-F | ATACGATTACTTTCTGTTCGAGGCCCCTCGGCTTGAACGAA |
| CDF-Cherry-R | GGTCTTGAGGGGTTTTTTGCGCAAGATCCGGCCACGATGA |
| (KKK) GetIFS-F | GTATAAGAAGGAGATATACATATGGCGAAGAAGAAGCTGAGCGCAAAAAGCAAAAG |
| GetIFS-R | CAGCGGTTTCTTTACCAGACTCGAGTTAAGAAGAAAACAGTTTCG |
| GmHID-F | CATGCCATGGCCAAAGAAATCGTTAAAG |
| GmHID-R | CCCAAGCTTTTAAACCAGAAAGCTGGCC |
| BglII-HID-F | CAGATCTCATGGCCAAAGAAATCGTTAAAGA |
| XhoI-IFS-R | GACTCGAGTTAAGAAGAAAACAGTTTCGG |
表2 本研究所用引物
Table 2 Primers used in this study
| Primers | Sequences(5'-3') |
|---|---|
| Cherry-CDF-F | GTCATCGTGGCCGGATCTTGCGCAAAAAACCCCTCAAGACC |
| Cherry-CDF-R | TTCGTTCAAGCCGAGGGGCCTCGAACAGAAAGTAATCGTA |
| CDF-Cherry-F | ATACGATTACTTTCTGTTCGAGGCCCCTCGGCTTGAACGAA |
| CDF-Cherry-R | GGTCTTGAGGGGTTTTTTGCGCAAGATCCGGCCACGATGA |
| (KKK) GetIFS-F | GTATAAGAAGGAGATATACATATGGCGAAGAAGAAGCTGAGCGCAAAAAGCAAAAG |
| GetIFS-R | CAGCGGTTTCTTTACCAGACTCGAGTTAAGAAGAAAACAGTTTCG |
| GmHID-F | CATGCCATGGCCAAAGAAATCGTTAAAG |
| GmHID-R | CCCAAGCTTTTAAACCAGAAAGCTGGCC |
| BglII-HID-F | CAGATCTCATGGCCAAAGAAATCGTTAAAGA |
| XhoI-IFS-R | GACTCGAGTTAAGAAGAAAACAGTTTCGG |

图2 构建LCA2G-LLQ4R共培养系统在M9Y培养基中从头合成甘草素(a) LLQ5单培养系统从头合成甘草素的动态曲线; (b) LCA2G-LLQ4R共培养系统的设计; (c) LCA2G-LLQ4R共培养系统接种比例优化
Fig.2 Engineering of LCA2G-LLQ4R coculture for de novo biosynthesis of liquiritigenin in M9Y medium
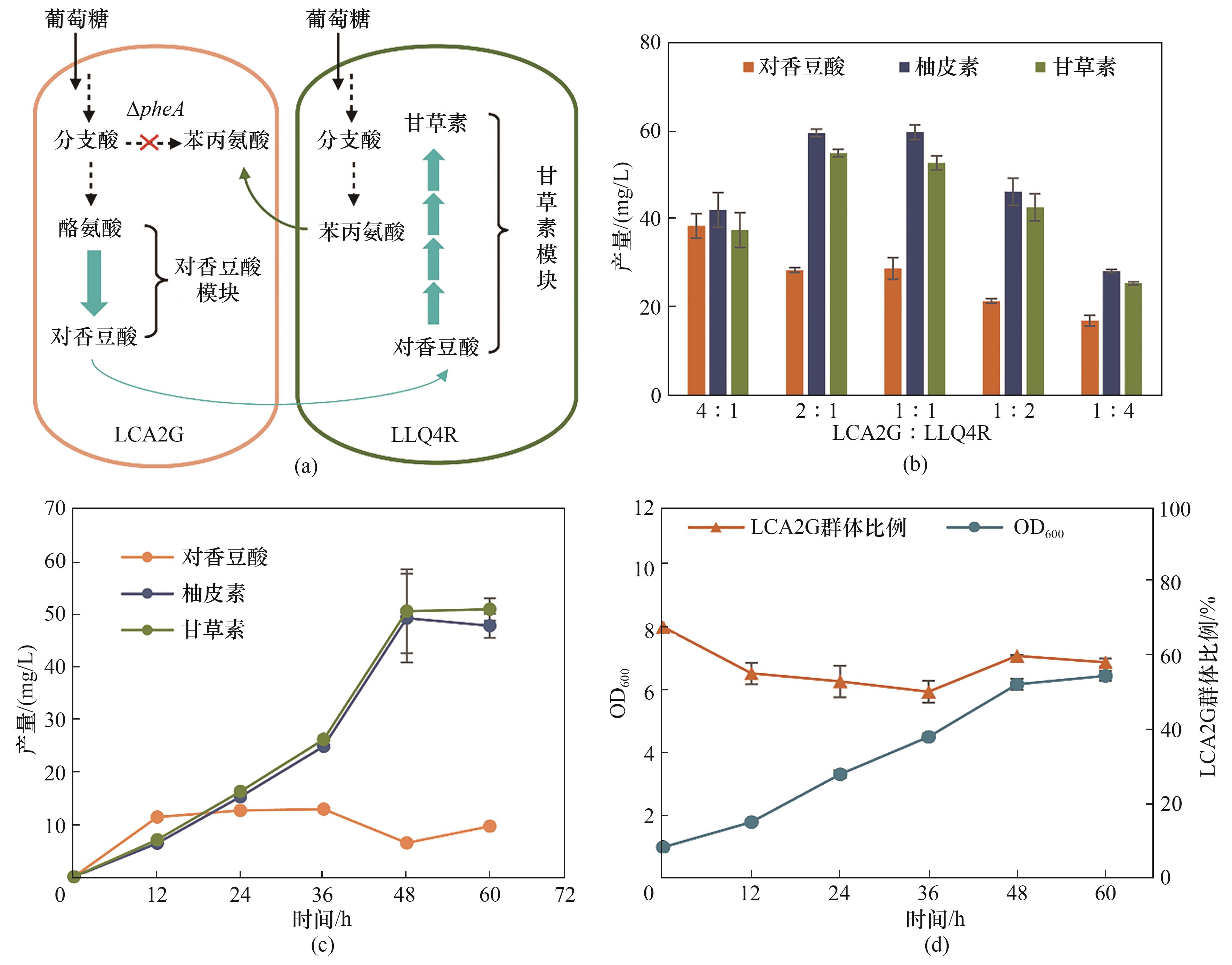
图3 构建LCA2G-LLQ4R偏利双菌共培养系统在M9培养基中从头合成甘草素(a) LCA2G-LLQ4R偏利共生共培养系统的设计; (b) LCA2G-LLQ4R共培养系统接种比例优化(发酵72 h); (c) 接种比例为2∶1时LCA2G-LLQ4R共培养系统中对香豆酸、柚皮素和甘草素产量的动力学曲线; (d) 接种比例为2∶1时LCA2G-LLQ4R共培养系统中生物量及LCA2G群体比例的动力学曲线
Fig.3 Commensalistic engineering of LCA2G-LLQ4R coculture for de novo biosynthesis of liquiritigenin in M9 medium
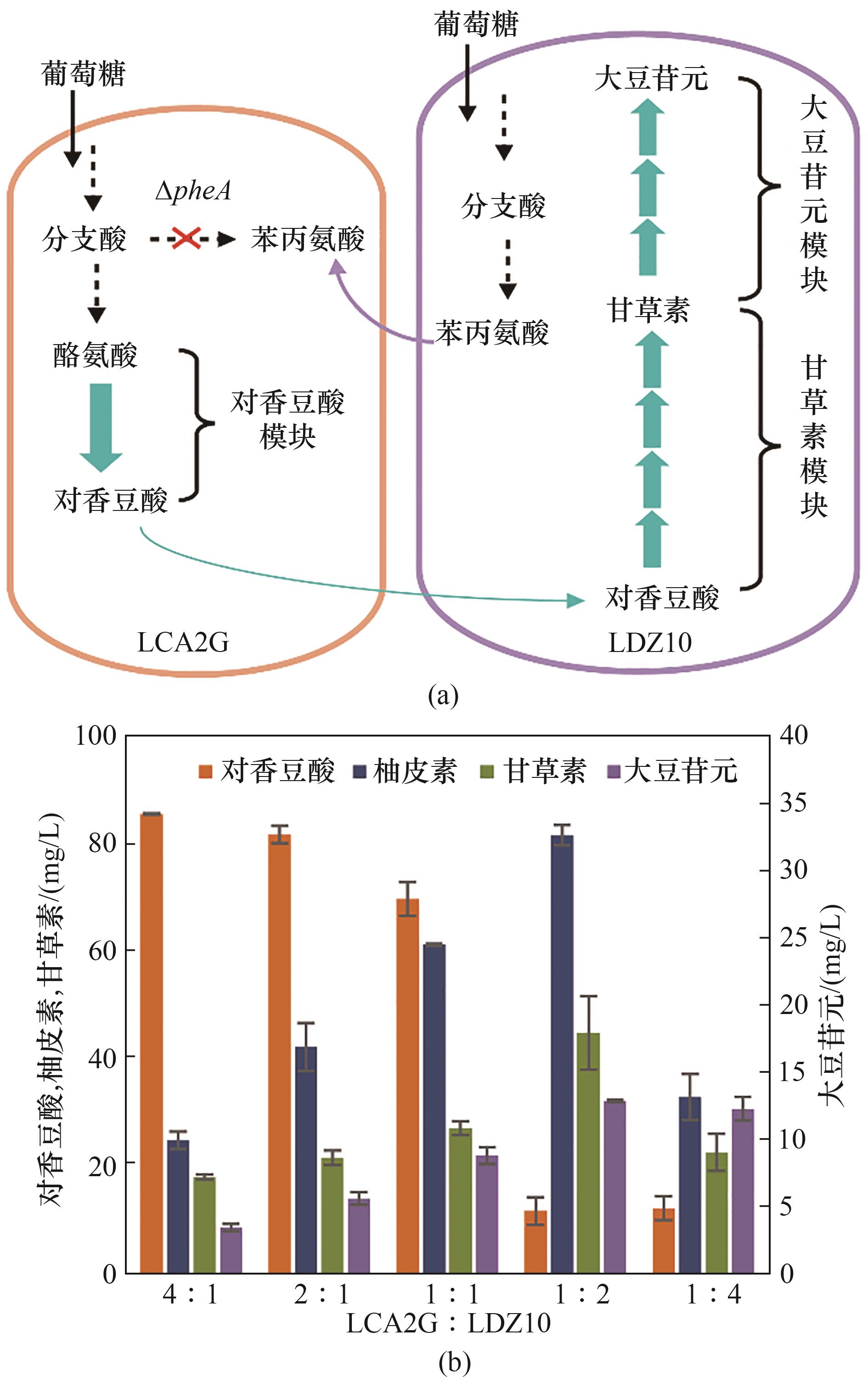
图4 构建LCA2G-LDZ10偏利双菌共培养系统在M9培养基中从头合成大豆苷元(a) LCA2G-LDZ10共培养系统的设计; (b) LCA2G-LDZ10共培养系统接种比例优化
Fig.4 Commensalistic engineering of LCA2G-LDZ10 coculture for de novo biosynthesis of daidzein in M9 medium
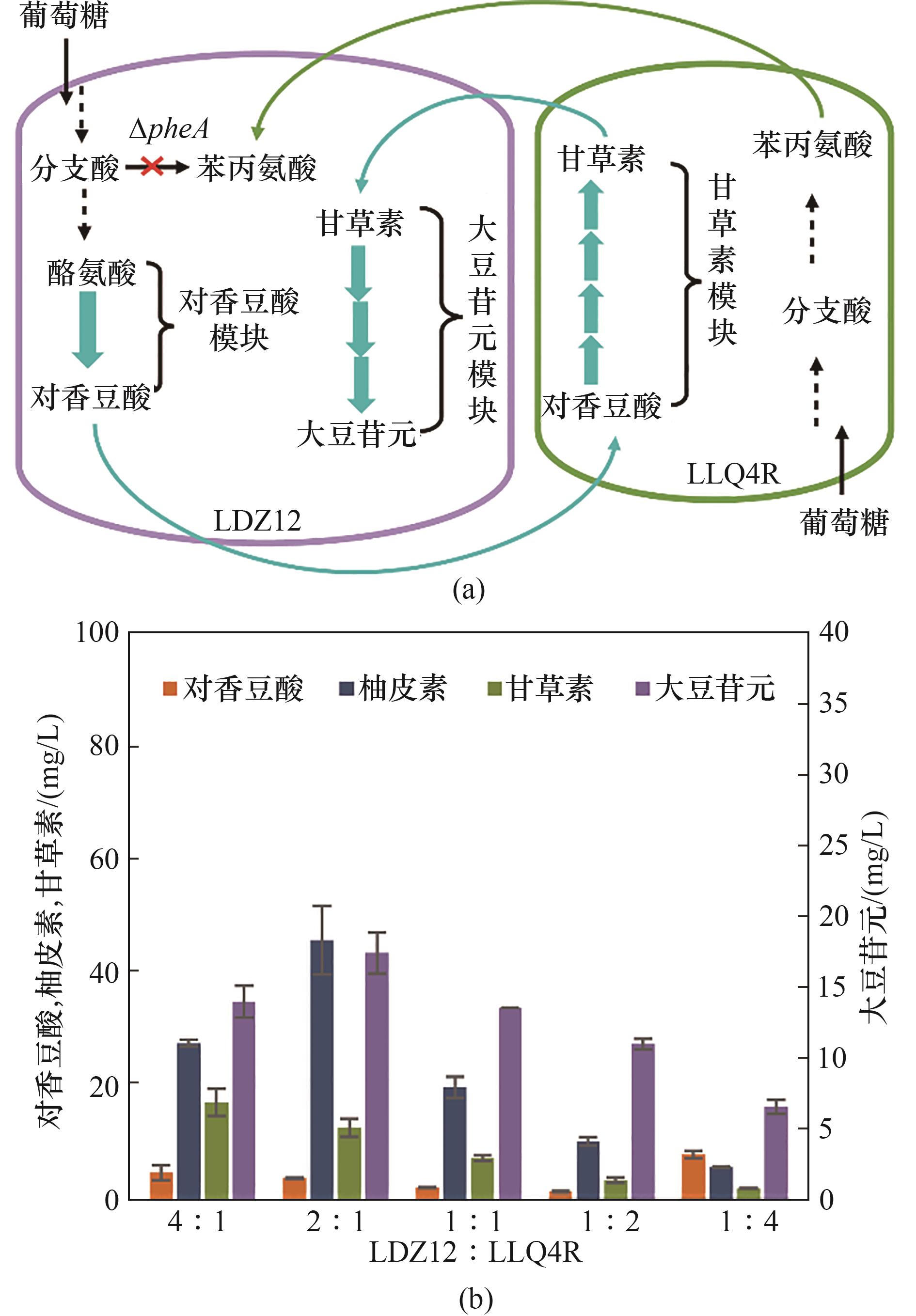
图5 构建LDZ12-LLQ4R偏利双菌共培养系统在M9培养基中从头合成大豆苷元(a) LDZ12-LLQ4R共培养系统的设计; (b) LDZ12-LLQ4R共培养系统接种比例优化
Fig.5 Commensalistic engineering of LDZ12-LLQ4R coculture for de novo biosynthesis of daidzein in M9 medium
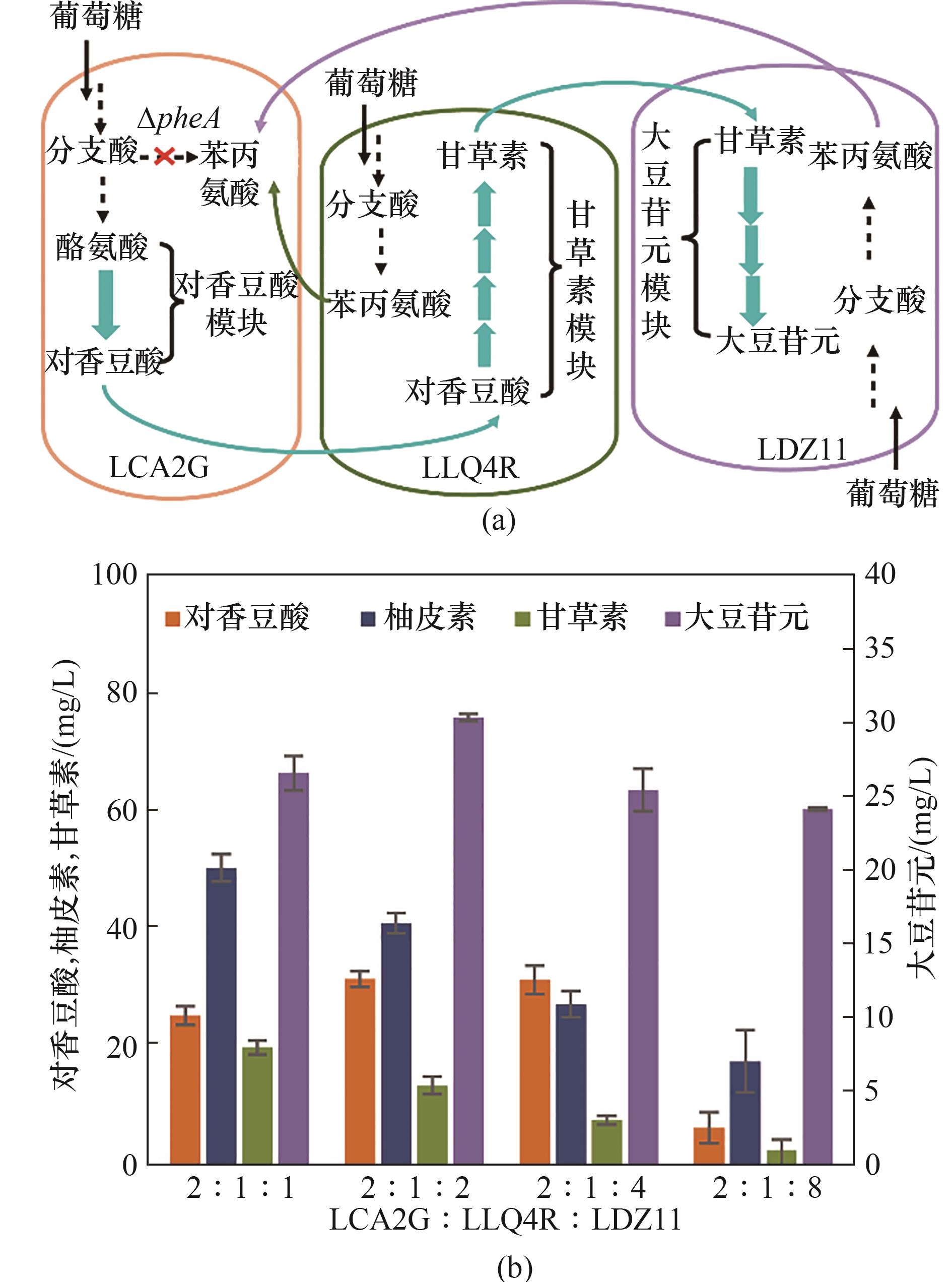
图6 构建LCA2G-LLQ4R-LDZ11偏利三菌共培养系统在M9培养基中从头合成大豆苷元(a) LCA2G-LLQ4R-LDZ11共培养系统的设计; (b) LCA2G-LLQ4R-LDZ11共培养系统接种比例优化
Fig.6 Commensalistic engineering of LCA2G-LLQ4R-LDZ11 coculture for de novo biosynthesis of daidzein in M9 medium

图7 LCA2G-LLQ4R-LDZ11偏利三菌共培养系统在M9培养基中合成大豆苷元的动态曲线(a) 接种比例2∶1∶2时LCA2G-LLQ4R-LDZ11共培养系统在M9培养基中合成的对香豆酸、柚皮素、甘草素和大豆苷元产量的动力学曲线; (b) 接种比例2∶1∶2时LCA2G-LLQ4R-LDZ11共培养系统在M9培养基中的生物量和群体比例的动力学曲线; (c) LDZ8单培养系统在M9Y培养基中合成大豆苷元
Fig.7 The time profile of commensalistic LCA2G-LLQ4R-LDZ11 coculture for biosynthesis of daidzein in M9 medium
| 1 | Křížová L, Dadáková K, Kašparovská J, et al. Isoflavones[J]. Molecules, 2019, 24(6): 1076. |
| 2 | Ahmed T, Javed S, Tariq A, et al. Daidzein and its effects on brain[J]. Current Medicinal Chemistry, 2017, 24(4): 365-375. |
| 3 | Meng H Z, Fu G H, Shen J, et al. Ameliorative effect of daidzein on cisplatin-induced nephrotoxicity in mice via modulation of inflammation, oxidative stress, and cell death[J]. Oxidative Medicine and Cellular Longevity, 2017, 2017: 3140680. |
| 4 | Yu Z Y, Yang L, Deng S, et al. Daidzein ameliorates LPS-induced hepatocyte injury by inhibiting inflammation and oxidative stress[J]. European Journal of Pharmacology, 2020, 885: 173399. |
| 5 | Blicharski T, Oniszczuk A. Extraction methods for the isolation of isoflavonoids from plant material[J]. Open Chemistry, 2017, 15(1): 34-45. |
| 6 | Sajid M, Stone S R, Kaur P. Recent advances in heterologous synthesis paving way for future green-modular bioindustries: a review with special reference to isoflavonoids[J]. Frontiers in Bioengineering and Biotechnology, 2021, 9: 673270. |
| 7 | Ajikumar P K, Xiao W H, Tyo K E J, et al. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli [J]. Science, 2010, 330(6000): 70-74. |
| 8 | Paddon C J, Westfall P J, Pitera D J, et al. High-level semi-synthetic production of the potent antimalarial artemisinin[J]. Nature, 2013, 496(7446): 528-532. |
| 9 | Waki T, Mameda R, Nakano T, et al. A conserved strategy of chalcone isomerase-like protein to rectify promiscuous chalcone synthase specificity[J]. Nature Communications, 2020, 11: 870. |
| 10 | Liu X, Li L L, Zhao G R. Systems metabolic engineering of Escherichia coli coculture for de novo production of genistein[J]. ACS Synthetic Biology, 2022, 11(5): 1746-1757. |
| 11 | Liu Q L, Liu Y, Li G, et al. De novo biosynthesis of bioactive isoflavonoids by engineered yeast cell factories[J]. Nature Communications, 2021, 12: 6085. |
| 12 | Stahlhut S G, Siedler S, Malla S, et al. Assembly of a novel biosynthetic pathway for production of the plant flavonoid fisetin in Escherichia coli [J]. Metabolic Engineering, 2015, 31: 84-93. |
| 13 | Yan Y J, Huang L X, Koffas M A G. Biosynthesis of 5-deoxyflavanones in microorganisms[J]. Biotechnology Journal, 2007, 2(10): 1250-1262. |
| 14 | Zhou K, Qiao K J, Edgar S, et al. Distributing a metabolic pathway among a microbial consortium enhances production of natural products[J]. Nature Biotechnology, 2015, 33(4): 377-383. |
| 15 | Chen Z Y, Sun X X, Li Y, et al. Metabolic engineering of Escherichia coli for microbial synthesis of monolignols[J]. Metabolic Engineering, 2017, 39: 102-109. |
| 16 | Jones J A, Wang X. Use of bacterial co-cultures for the efficient production of chemicals[J]. Current Opinion in Biotechnology, 2018, 53: 33-38. |
| 17 | Sgobba E, Stumpf A K, Vortmann M, et al. Synthetic Escherichia coli-Corynebacterium glutamicum consortia for L-lysine production from starch and sucrose[J]. Bioresource Technology, 2018, 260: 302-310. |
| 18 | Li S Z, Liang C N, Liu G X, et al. De novo biosynthesis of chlorogenic acid using an artificial microbial community[J]. Journal of Agricultural and Food Chemistry, 2021, 69(9): 2816-2825. |
| 19 | Liu X, Liu J C, Lei D W, et al. Modular metabolic engineering for production of phloretic acid, phloretin and phlorizin in Escherichia coli [J]. Chemical Engineering Science, 2022, 247: 116931. |
| 20 | Yao Y F, Wang C S, Qiao J J, et al. Metabolic engineering of Escherichia coli for production of salvianic acid A via an artificial biosynthetic pathway[J]. Metabolic Engineering, 2013, 19: 79-87. |
| 21 | Rodriguez A, Strucko T, Stahlhut S G, et al. Metabolic engineering of yeast for fermentative production of flavonoids[J]. Bioresource Technology, 2017, 245: 1645-1654. |
| 22 | Akram M, Rasool A, An T, et al. Metabolic engineering of Yarrowia lipolytica for liquiritigenin production[J]. Chemical Engineering Science, 2021, 230: 116177. |
| 23 | Santos C N S, Koffas M, Stephanopoulos G. Optimization of a heterologous pathway for the production of flavonoids from glucose[J]. Metabolic Engineering, 2011, 13(4): 392-400. |
| 24 | Chemler J A, Lim C G, Daiss J L, et al. A versatile microbial system for biosynthesis of novel polyphenols with altered estrogen receptor binding activity[J]. Chemistry & Biology, 2010, 17(4): 392-401. |
| 25 | Wang X N, Li Z H, Policarpio L, et al. De novo biosynthesis of complex natural product sakuranetin using modular co-culture engineering[J]. Applied Microbiology and Biotechnology, 2020, 104(11): 4849-4861. |
| 26 | Qiu Z T, Liu X, Li J, et al. Metabolic division in an Escherichia coli coculture system for efficient production of kaempferide[J]. ACS Synthetic Biology, 2022, 11(3): 1213-1227. |
| 27 | Hom E F Y, Murray A W. Niche engineering demonstrates a latent capacity for fungal-algal mutualism[J]. Science, 2014, 345(6192): 94-98. |
| 28 | Li Z H, Wang X N, Zhang H R. Balancing the non-linear rosmarinic acid biosynthetic pathway by modular co-culture engineering[J]. Metabolic Engineering, 2019, 54: 1-11. |
| 29 | Li X L, Zhou Z, Li W N, et al. Design of stable and self-regulated microbial consortia for chemical synthesis[J]. Nature Communications, 2022, 13: 1554. |
| 30 | Jones J A, Vernacchio V R, Collins S M, et al. Complete biosynthesis of anthocyanins using E. coli polycultures[J]. mBio, 2017, 8(3): e00621-e00617. |
| 31 | Wu S, Ma X Q, Zhou A Q, et al. Establishment of strigolactone-producing bacterium-yeast consortium[J]. Science Advances, 2021, 7(38): eabh4048. |
| 32 | Wu S B, Xue Y T, Yang S J, et al. Combinational quorum sensing devices for dynamic control in cross-feeding cocultivation[J]. Metabolic Engineering, 2021, 67: 186-197. |
| 33 | Mameda R, Waki T, Kawai Y, et al. Involvement of chalcone reductase in the soybean isoflavone metabolon: identification of GmCHR5, which interacts with 2-hydroxyisoflavanone synthase[J]. The Plant Journal, 2018, 96(1): 56-74. |
| 34 | Gul K, Singh A K, Jabeen R. Nutraceuticals and functional foods: the foods for the future world[J]. Critical Reviews in Food Science and Nutrition, 2016, 56(16): 2617-2627. |
| 35 | Wang X, Li C F, Zhou C, et al. Molecular characterization of the C-glucosylation for puerarin biosynthesis in Pueraria lobata [J]. The Plant Journal, 2017, 90(3): 535-546. |
| 36 | Liu X, Li X B, Jiang J L, et al. Convergent engineering of syntrophic Escherichia coli coculture for efficient production of glycosides[J]. Metabolic Engineering, 2018, 47: 243-253. |
| 37 | Zhang H R, Wang X N. Modular co-culture engineering, a new approach for metabolic engineering[J]. Metabolic Engineering, 2016, 37: 114-121. |
| [1] | 赵春雷, 郭亮, 高聪, 宋伟, 吴静, 刘佳, 刘立明, 陈修来. 代谢工程改造大肠杆菌生产软骨素[J]. 化工学报, 2023, 74(5): 2111-2122. |
| [2] | 刘昕, 戈钧, 李春. 光驱动微生物杂合系统提高生物制造水平[J]. 化工学报, 2023, 74(1): 330-341. |
| [3] | 孙怡, 张腾, 吕波, 李春. 胞内生物传感器提高微生物细胞工厂的精细调控[J]. 化工学报, 2022, 73(2): 521-534. |
| [4] | 王靖楠, 庞建, 秦磊, 郭超, 吕波, 李春, 王超. 丁烯基多杀菌素高产菌株的选育和改造策略[J]. 化工学报, 2022, 73(2): 566-576. |
| [5] | 王欣慧, 王颖, 姚明东, 肖文海. 维生素A生物合成的研究进展[J]. 化工学报, 2022, 73(10): 4311-4323. |
| [6] | 周武林, 高惠芳, 吴玉玲, 张显, 徐美娟, 杨套伟, 邵明龙, 饶志明. 重组酿酒酵母生物合成菜油甾醇[J]. 化工学报, 2021, 72(8): 4314-4324. |
| [7] | 王欣, 赵鹏, 李清扬, 田平芳. 半导体合成生物学的研究进展[J]. 化工学报, 2021, 72(5): 2426-2435. |
| [8] | 毛金竹, 肖淑玲, 杨智淳, 王孝宇, 张诗, 陈俊宏, 谢佶晟, 陈福德, 黄子诺, 冯天宇, 张瑷珲, 方柏山. 合成生物学在农残检测领域的应用[J]. 化工学报, 2021, 72(5): 2413-2425. |
| [9] | 郑煜堃, 孙青, 陈振, 于慧敏. 微生物细胞工厂生产化学品的研究进展——以几种典型小分子和大分子化学品为例[J]. 化工学报, 2021, 72(12): 6109-6121. |
| [10] | 赵贞尧, 张保财, 李锋, 宋浩. 产电细胞的合成生物学设计构建[J]. 化工学报, 2021, 72(1): 468-482. |
| [11] | 王炼, 吴迪, 周景文. 木脂素的生物合成及其微生物法生产的研究进展[J]. 化工学报, 2021, 72(1): 320-333. |
| [12] | 王凯峰, 王金鹏, 韦萍, 纪晓俊. 代谢工程改造解脂耶氏酵母生产脂肪酸及其衍生物[J]. 化工学报, 2021, 72(1): 351-365. |
| [13] | 高虎涛, 申晓林, 孙新晓, 王佳, 袁其朋. 代谢工程调控策略在生物合成氨基酸及其衍生物中的应用[J]. 化工学报, 2020, 71(9): 4058-4070. |
| [14] | 徐静, 由紫暄, 张君奇, 陈正, 吴德光, 李锋, 宋浩. 合成生物学方法改造电活性生物膜研究进展[J]. 化工学报, 2020, 71(9): 3950-3962. |
| [15] | 秦磊, 俞杰, 宁小钰, 孙文涛, 李春. 合成生物系统构建与绿色生物“智”造[J]. 化工学报, 2020, 71(9): 3979-3994. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备 11010102001995号
京公网安备 11010102001995号