化工学报 ›› 2022, Vol. 73 ›› Issue (2): 566-576.DOI: 10.11949/0438-1157.20210898
王靖楠1( ),庞建2,秦磊2,郭超3,吕波1,李春1,2(
),庞建2,秦磊2,郭超3,吕波1,李春1,2( ),王超3(
),王超3( )
)
收稿日期:2021-06-30
修回日期:2021-11-01
出版日期:2022-02-05
发布日期:2022-02-18
通讯作者:
李春,王超
作者简介:王靖楠(1997—),女,硕士研究生,基金资助:
Jingnan WANG1( ),Jian PANG2,Lei QIN2,Chao GUO3,Bo LYU1,Chun LI1,2(
),Jian PANG2,Lei QIN2,Chao GUO3,Bo LYU1,Chun LI1,2( ),Chao WANG3(
),Chao WANG3( )
)
Received:2021-06-30
Revised:2021-11-01
Online:2022-02-05
Published:2022-02-18
Contact:
Chun LI,Chao WANG
摘要:
丁烯基多杀菌素(butenyl-spinosyn)是一种由须糖多孢菌(Saccharopolyspora pogona)产生的杀虫剂,兼具生物农药的安全性和化学农药的速效性。当前,野生型须糖多孢菌合成丁烯基多杀菌素效率低,达不到工业生产要求,获得高产菌株是亟待解决的问题。目前关于丁烯基多杀菌素的相关研究较少,由刺糖多孢菌(Saccharopolyspora spinosa)产生的多杀菌素是丁烯基多杀菌素的结构类似物,具有相似的生物合成途径,本文介绍了两者的基本特性,借鉴多杀菌素的研究经验总结了丁烯基多杀菌素高产菌株已有及可用的选育和改造策略,包括传统的理化诱变方法以及代谢流调控、途径基因调控、转录调控、异源表达等更加精准的基因工程方法,以期为后续丁烯基多杀菌素的深入研究提供思路。
中图分类号:
王靖楠, 庞建, 秦磊, 郭超, 吕波, 李春, 王超. 丁烯基多杀菌素高产菌株的选育和改造策略[J]. 化工学报, 2022, 73(2): 566-576.
Jingnan WANG, Jian PANG, Lei QIN, Chao GUO, Bo LYU, Chun LI, Chao WANG. Breeding and modification strategies of butenyl-spinosyn high-yield strains[J]. CIESC Journal, 2022, 73(2): 566-576.
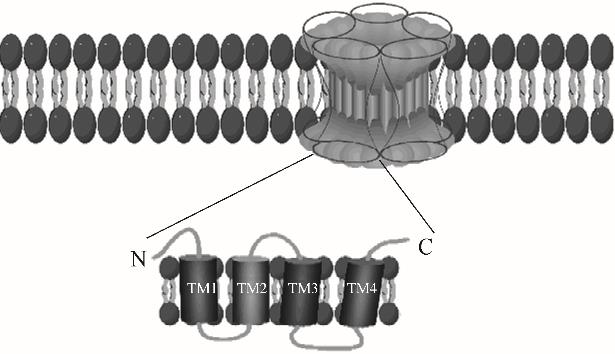
图2 nAChRs示意图[8]nAChRs是动物的主要兴奋性神经递质受体,由五个跨膜亚基组成,它们围绕着一充满水的中心孔。每个亚单位包括一个参与激动剂结合的N-末端胞外区域、四个跨膜区域(TM1~TM4)和一个C-末端胞外区域
Fig.2 Schematic diagram of nAChRs[8]
| 分类 | 诱变技术 | 机理[ | 产量变化 |
|---|---|---|---|
| 物理诱变 | UV诱变 | 使嘧啶形成二聚体 | 4.84倍,从77 mg/L到373 mg/L[ |
| 60Co诱变 | 引起DNA单链或双链断裂 | 1.16倍,从404 mg/L到469 mg/L[ | |
| 化学诱变 | NTG诱变 | 烷基取代核酸碱基氢原子引起碱基错配[ | 1.44倍,从436 mg/L到628 mg/L[ |
| MNNG诱变 | 2.28倍,从102 mg/L到232 mg/L[ | ||
| 生物诱变 | 基因组重排 | 通过原生质体融合富集理化诱变得到的正向 突变 | 6.60倍,从50 mg/L到332 mg/L[ |
| 转座诱变 | 转座子片段随机插入到基因组产生突变 | 1.14倍,从129 mg/L到147 mg/L[ | |
| 新型诱变技术 | ARTP诱变 | 激发态的自由基等活性粒子损伤DNA | 1.32倍,从417 mg/L到550 mg/L[ |
| 航空搭载育种 | 太空环境的微重力等条件可能影响DNA | 3.88倍,从38 mg/L到148 mg/L[ | |
| 核糖体工程 | 核糖体蛋白突变影响次级代谢产物产生 | 1.24倍[ | |
| 复合诱变 | MNNG、60Co诱变 | 增加突变概率 | 1.87倍,从549 mg/L到1035 mg/L[ |
| EMS及ARTP诱变、链霉素诱变 | 增加突变概率 | 7.71倍,从158 mg/L到1218 mg/L[ | |
| N+离子注入诱变、鼠李糖及多杀菌素 耐受 | 能量沉积、粒子注入、动量传递等过程与生物 体发生作用 | 1.44倍,从651 mg/L 到937 mg/L[ | |
| MPMS、核糖体工程 | 等离子体中的活性成分损伤生物大分子, 微生物产生大量随机突变[ | 1.29倍[ |
表1 多杀菌素高产菌株的诱变选育方法
Table 1 Mutation breeding method to high-yield strains of spinosyn
| 分类 | 诱变技术 | 机理[ | 产量变化 |
|---|---|---|---|
| 物理诱变 | UV诱变 | 使嘧啶形成二聚体 | 4.84倍,从77 mg/L到373 mg/L[ |
| 60Co诱变 | 引起DNA单链或双链断裂 | 1.16倍,从404 mg/L到469 mg/L[ | |
| 化学诱变 | NTG诱变 | 烷基取代核酸碱基氢原子引起碱基错配[ | 1.44倍,从436 mg/L到628 mg/L[ |
| MNNG诱变 | 2.28倍,从102 mg/L到232 mg/L[ | ||
| 生物诱变 | 基因组重排 | 通过原生质体融合富集理化诱变得到的正向 突变 | 6.60倍,从50 mg/L到332 mg/L[ |
| 转座诱变 | 转座子片段随机插入到基因组产生突变 | 1.14倍,从129 mg/L到147 mg/L[ | |
| 新型诱变技术 | ARTP诱变 | 激发态的自由基等活性粒子损伤DNA | 1.32倍,从417 mg/L到550 mg/L[ |
| 航空搭载育种 | 太空环境的微重力等条件可能影响DNA | 3.88倍,从38 mg/L到148 mg/L[ | |
| 核糖体工程 | 核糖体蛋白突变影响次级代谢产物产生 | 1.24倍[ | |
| 复合诱变 | MNNG、60Co诱变 | 增加突变概率 | 1.87倍,从549 mg/L到1035 mg/L[ |
| EMS及ARTP诱变、链霉素诱变 | 增加突变概率 | 7.71倍,从158 mg/L到1218 mg/L[ | |
| N+离子注入诱变、鼠李糖及多杀菌素 耐受 | 能量沉积、粒子注入、动量传递等过程与生物 体发生作用 | 1.44倍,从651 mg/L 到937 mg/L[ | |
| MPMS、核糖体工程 | 等离子体中的活性成分损伤生物大分子, 微生物产生大量随机突变[ | 1.29倍[ |
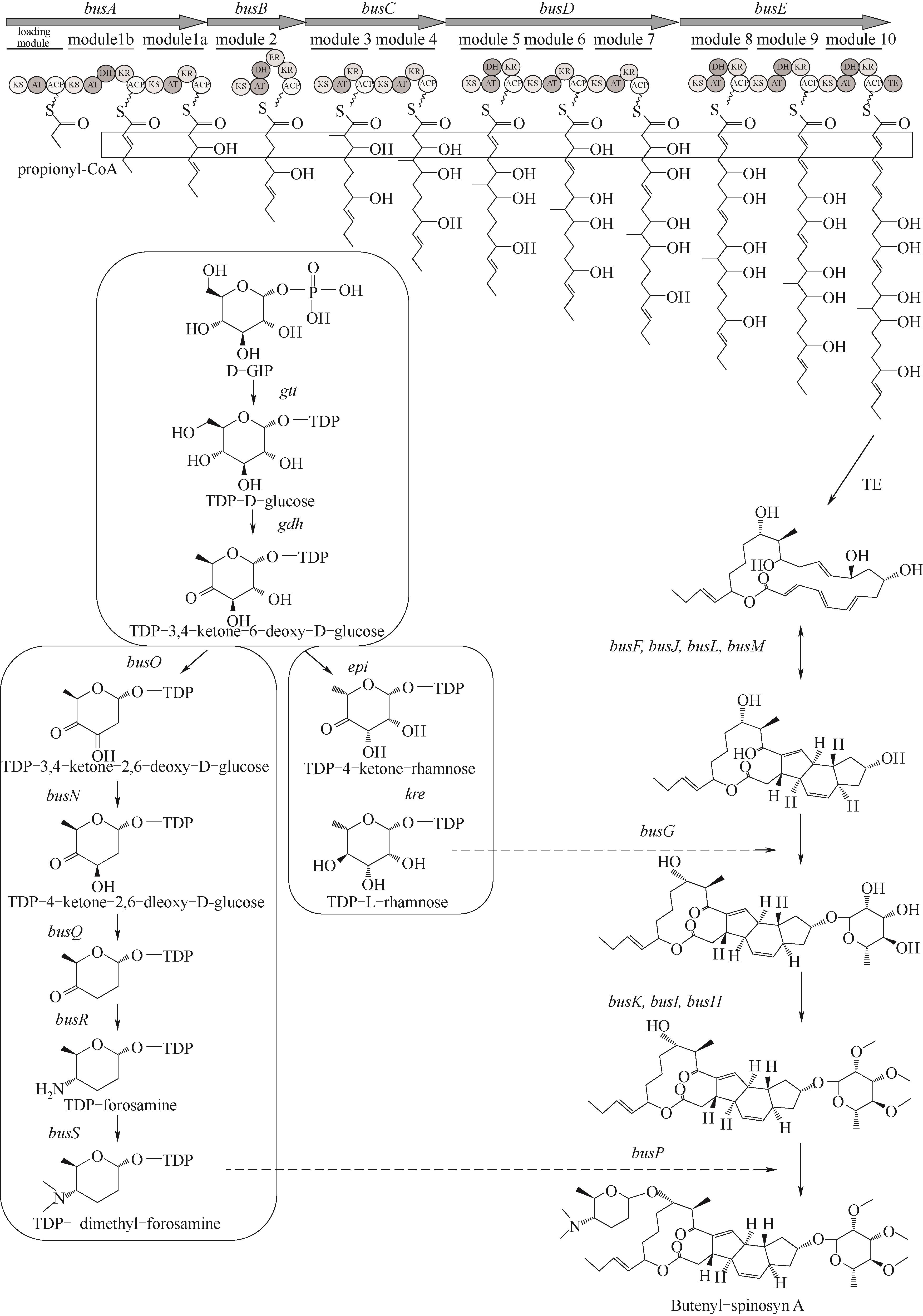
图4 丁烯基多杀菌素生物合成途径busA~E—聚酮合酶(PKS)基因;busJ、busF、busL、busM—交联桥基因;gtt—葡萄糖核苷转移酶基因;gdh—葡萄糖脱氢酶基因;epi—3', 5'-差向异构酶基因;kre—4'-酮还原酶基因;busG—鼠李糖转移酶基因;busH、busI、busK—氧甲基转移酶基因;busO—2, 3-脱氢酶基因;busN—3-酮还原酶基因;busQ—3, 4-脱氢酶基因;busR—转氨酶基因;busS—二甲基转移酶基因;busP—福乐糖胺转移酶基团;ACP—酰基载体蛋白;AT—酰基转移酶;KS—β-酮酰合酶;KR—β-酮酰还原酶;DH—β-羟酰脱水酶;ER—烯酰还原酶;TE—硫脂酶[3]
Fig.4 Biosynthesis pathway of butenyl-spinosyn
| 调控 | 描述 | 基因编辑方法 | 产量变化 | 分析机理 |
|---|---|---|---|---|
| 过表达PNPase[ | 转录因子,多核苷酸磷酸化酶,与氨基酸代谢、有机酸代谢及细胞生物合成相关酶等相关 | 同源重组- 单交换 | 1.96倍 | 减少菌丝体聚集,增加孢子产生,影响核苷酸代谢从而影响能量供应 |
| 过表达AfsR[ | 与链霉菌AfsR同源,属于抗生素调节蛋白家族 | 同源重组- 单交换 | 1.17倍 | 减少菌丝体聚集,影响产孢,降低初级代谢提升次级代谢 |
| 敲除padR[ | 普遍的调控因子,在链霉菌中与抗生素生物合成相关 | 同源重组- 双交换 | 1.27倍 | 促进转运相关蛋白的表达 |
| 过表达Sp1418[ | TetR家族的转录调控因子,与营养生长、菌丝分化及氧化应激相关 | 同源重组- 双交换 | 2.50倍 | 影响菌体氧化应激 |
| 过表达regX3[ | Sen X3-Reg X3双组分系统组分,与无机磷吸收相关 | 同源重组- 单交换 | 2.30倍 | 全局调控因子,无磷条件培养使得磷吸收最少 |
| 敲除lytS-L[ | LytTR家族双组分系统组分,传感器激酶基因,与改善营养环境相关 | 同源重组- 单交换 | 0.60倍 | 全局调控因子 |
| 敲除SP_1288[ | TetR家族调控蛋白 | CRISPR-Cas9 | 3.10倍 | 全局调控因子 |
表2 丁烯基多杀菌素转录调控方法提产
Table 2 Methods of improving yield by genetic engineering of butenyl spinosyn
| 调控 | 描述 | 基因编辑方法 | 产量变化 | 分析机理 |
|---|---|---|---|---|
| 过表达PNPase[ | 转录因子,多核苷酸磷酸化酶,与氨基酸代谢、有机酸代谢及细胞生物合成相关酶等相关 | 同源重组- 单交换 | 1.96倍 | 减少菌丝体聚集,增加孢子产生,影响核苷酸代谢从而影响能量供应 |
| 过表达AfsR[ | 与链霉菌AfsR同源,属于抗生素调节蛋白家族 | 同源重组- 单交换 | 1.17倍 | 减少菌丝体聚集,影响产孢,降低初级代谢提升次级代谢 |
| 敲除padR[ | 普遍的调控因子,在链霉菌中与抗生素生物合成相关 | 同源重组- 双交换 | 1.27倍 | 促进转运相关蛋白的表达 |
| 过表达Sp1418[ | TetR家族的转录调控因子,与营养生长、菌丝分化及氧化应激相关 | 同源重组- 双交换 | 2.50倍 | 影响菌体氧化应激 |
| 过表达regX3[ | Sen X3-Reg X3双组分系统组分,与无机磷吸收相关 | 同源重组- 单交换 | 2.30倍 | 全局调控因子,无磷条件培养使得磷吸收最少 |
| 敲除lytS-L[ | LytTR家族双组分系统组分,传感器激酶基因,与改善营养环境相关 | 同源重组- 单交换 | 0.60倍 | 全局调控因子 |
| 敲除SP_1288[ | TetR家族调控蛋白 | CRISPR-Cas9 | 3.10倍 | 全局调控因子 |
| 宿主菌株 | 提产方法 | 产量提升 | 基因簇克隆方法 |
|---|---|---|---|
| S. albus J1074[ | 1.组学分析找到三个限速问题:鼠李糖生物合成不足、 甲基转移酶活性不足以及聚酮合酶活性不足; 2.改进上述限速步骤 | 到1.4 mg/L | BAC文库 |
| S. coelicolor M145[ | 过表达鼠李糖合成基因 | 到1.0 mg/L; 到1.5 mg/L | BAC文库 |
| S. albus J1074[ | 1.构建79 kb的人工基因簇,将通路分成7个操纵子; 2.每个操纵子都以一个强组成性启动子表达 | 到1.1 mg/L | ExoCET[ |
| Sa. erythraea[ | 1.替换红色糖多孢菌的PKS基因簇; 2.将spnGF替换为eryAB; 3.引入sfp基因; 4.过表达鼠李糖合成基因 | 到830.0 mg/L | Cosmid文库 |
表3 多杀菌素的异源合成
Table 3 Heterologous synthesis of spinosyn
| 宿主菌株 | 提产方法 | 产量提升 | 基因簇克隆方法 |
|---|---|---|---|
| S. albus J1074[ | 1.组学分析找到三个限速问题:鼠李糖生物合成不足、 甲基转移酶活性不足以及聚酮合酶活性不足; 2.改进上述限速步骤 | 到1.4 mg/L | BAC文库 |
| S. coelicolor M145[ | 过表达鼠李糖合成基因 | 到1.0 mg/L; 到1.5 mg/L | BAC文库 |
| S. albus J1074[ | 1.构建79 kb的人工基因簇,将通路分成7个操纵子; 2.每个操纵子都以一个强组成性启动子表达 | 到1.1 mg/L | ExoCET[ |
| Sa. erythraea[ | 1.替换红色糖多孢菌的PKS基因簇; 2.将spnGF替换为eryAB; 3.引入sfp基因; 4.过表达鼠李糖合成基因 | 到830.0 mg/L | Cosmid文库 |
| 69 | Huang J, Yu Z, Li M H, et al. High level of spinosad production in the heterologous host Saccharopolyspora erythraea[J]. Applied and Environmental Microbiology, 2016, 82(18): 5603-5611. |
| 70 | Zhang X F, Hindra, Elliot M A. Unlocking the trove of metabolic treasures: activating silent biosynthetic gene clusters in bacteria and fungi[J]. Current Opinion in Microbiology, 2019, 51: 9-15. |
| 1 | 史雪岩. 多杀菌素类杀虫剂的环境降解及抗性机制研究进展[J]. 农药学学报, 2018, 20(5): 557-567. |
| Shi X Y. Research progresses on environmental degradation and resistance mechanism of spinosyn insecticides[J]. Chinese Journal of Pesticide Science, 2018, 20(5): 557-567. | |
| 2 | Zhang K, Li J R, Wen D S, et al. Study on the synthesis and insecticidal activity of spinosyn a derivatives[J]. Chinese Journal of Organic Chemistry, 2018, 38(12): 3363. |
| 3 | 寿佳丽, 裘娟萍. 新型生物农药——丁烯基多杀菌素[J]. 农药, 2011, 50(4): 239-243, 272. |
| Shou J L, Qiu J P. A new type of biological pesticide—butenyl-spinosyns[J]. Agrochemicals, 2011, 50(4): 239-243, 272. | |
| 4 | Whitman W B. Bergey's Manual of Systematic Bacteriology: Volume 5[M]. 2nd ed. USA, 2012: 1396-1415. |
| 5 | 郭超, 赵晨, 黎琪, 等. 产丁烯基多杀菌素菌株的筛选及鉴定[J]. 粮油食品科技, 2019, 27(2): 55-60. |
| Guo C, Zhao C, Li Q, et al. Screening and identification of the strain producing butenyl-spinosyns[J]. Science and Technology of Cereals, Oils and Foods, 2019, 27(2): 55-60. | |
| 6 | APRD. Arthropod Pesticide Resistance Database[DB/OL]. [2021-06-23]. |
| 7 | Yin X H, Wu Q J, Zhang Y J, et al. Analysis of persistent changes to γ-aminobutyric acid receptor gene expression in Plutella xylostella subjected to sublethal amounts of spinosad[J]. Genetics and Molecular Research, 2016, 15(3): gmr.15038782. |
| 71 | Sparks T C, Crouse G D, Benko Z, et al. The spinosyns, spinosad, spinetoram, and synthetic spinosyn mimics — discovery, exploration, and evolution of a natural product chemistry and the impact of computational tools[J]. Pest Management Science, 2021, 77(8): 3637-3649. |
| 8 | Thany S H, Lenaers G, Raymond-Delpech V, et al. Exploring the pharmacological properties of insect nicotinic acetylcholine receptors[J]. Trends in Pharmacological Sciences, 2007, 28(1): 14-22. |
| 9 | Guillem-Amat A, Sánchez L, López-Errasquín E, et al. Field detection and predicted evolution of spinosad resistance in Ceratitis capitata[J]. Pest Management Science, 2020, 76(11): 3702-3710. |
| 10 | Santos V S V, Pereira B B. Properties, toxicity and current applications of the biolarvicide spinosad[J]. Journal of Toxicology and Environmental Health, Part B, 2020, 23(1): 13-26. |
| 11 | Zhang Y, Guo W, Chen H, et al. Spinetoram confers its cytotoxic effects by inducing AMPK/mTOR-mediated autophagy and oxidative DNA damage[J]. Ecotoxicology and Environmental Safety, 2019, 183: 109480. |
| 12 | 袁姚梦, 邢新会, 张翀. 微生物细胞工厂的设计构建: 从诱变育种到全基因组定制化创制[J]. 合成生物学, 2020, 1(6): 656-673. |
| Yuan Y M, Xing X H, Zhang C. Progress and prospective of engineering microbial cell factories: from random mutagenesis to customized design in genome scale[J]. Synthetic Biology Journal, 2020, 1(6): 656-673. | |
| 13 | Jha A K, Pokhrel A R, Chaudhary A K, et al. Metabolic engineering of rational screened Saccharopolyspora spinosa for the enhancement of spinosyns A and D production[J]. Molecules and Cells, 2014, 37(10): 727-733. |
| 14 | 代鹏, 徐雪莲, 贺玉平, 等. 多杀菌素生产菌株的选育[J]. 热带作物学报, 2005, 26(4): 67-70. |
| Dai P, Xu X L, He Y P, et al. Breeding of production strains of spinosads[J]. Chinese Journal of Tropical Crops, 2005, 26(4): 67-70. | |
| 15 | 宋炜, 熊犍, 郭伟群, 等. MNNG对多杀菌素产生菌的诱变效应[J]. 中国生物防治, 2009, 25(2): 176-180. |
| Song W, Xiong J, Guo W Q, et al. Mutagenic effects of MNNG treatment on the spinosad producing strains[J]. Chinese Journal of Biological Control, 2009, 25(2): 176-180. | |
| 16 | 陈园. 多杀菌素高产菌株的诱变选育及快速筛选方法的研究[D]. 广州: 华南理工大学, 2013. |
| Chen Y. Mutation breeding of spinosad high-producing strain and study of rapid screening method[D]. Guangzhou: South China University of Technology, 2013. | |
| 17 | Wang H, Xue W, He Y M, et al. Improvement of the ability to produce spinosad in Saccharopolyspora spinosa through the acquisition of drug resistance and genome shuffling[J]. Annals of Microbiology, 2015, 65(2): 771-777. |
| 18 | 白露露, 贺卫军, 龙青山, 等. 糖多孢菌属菌株的体内转座诱变系统[J]. 华中农业大学学报, 2019, 38(5): 85-91. |
| Bai L L, He W J, Long Q S, et al. In vivo transposition mutagenesis system of Saccharopolyspora[J]. Journal of Huazhong Agricultural University, 2019, 38(5): 85-91. | |
| 19 | 陈继红, 张利平, 郭立格, 等. 多杀菌素产生菌株航天育种效果研究[J]. 安徽农业科学, 2008, 36(12): 4951-4952, 4957. |
| Chen J H, Zhang L P, Guo L G, et al. Breeding effect of space treatment breeding on producting strains of spinosad[J]. Journal of Anhui Agricultural Sciences, 2008, 36(12): 4951-4952, 4957. | |
| 20 | 王海霞, 陈园, 王超, 等. 组合抗生素抗性选育多杀菌素高产菌株[J]. 粮油食品科技, 2017, 25(4): 70-75. |
| Wang H X, Chen Y, Wang C, et al. Breeding of spinosad high-producing strains by combinatorial antibiotic resistance[J]. Science and Technology of Cereals, Oils and Foods, 2017, 25(4): 70-75. | |
| 21 | Yang G J, He Y P, Jiang Y, et al. A new medium for improving spinosad production by Saccharopolyspora spinosa[J]. Jundishapur Journal of Microbiology, 2016, 9(6): e16765. |
| 22 | 王欣荣, 张爱雪, 唐智超, 等. 产多杀菌素刺糖多孢菌的诱变选育[J]. 中国抗生素杂志, 2020, 45(9): 873-877. |
| Wang X R, Zhang A X, Tang Z C, et al. Mutagenesis and breeding of spinosad-producing strains of Saccharopolysporaspinosa[J]. Chinese Journal of Antibiotics, 2020, 45(9): 873-877. | |
| 23 | 郭伟群, 罗莉斯, 李能威, 等. 利用离子注入技术选育多杀菌素高产菌株[J]. 中国农业科技导报, 2012, 14(4): 148-152. |
| Guo W Q, Luo L S, Li N W, et al. Breeding high-yield spinosad-producing strain by nitrogen ion implantation[J]. Journal of Agricultural Science and Technology, 2012, 14(4): 148-152. | |
| 24 | 王海霞, 陈园, 王超, 等. MPMS诱变结合抗生素抗性选育多杀菌素高产菌株[J]. 粮油食品科技, 2017, 25(3): 82-86. |
| Wang H X, Chen Y, Wang C, et al. Breeding of spinosad high-producing strains through MPMS mutagenesis combined with antibiotics[J]. Science and Technology of Cereals, Oils and Foods, 2017, 25(3): 82-86. | |
| 25 | Li Y Y, Chang X, Yu W B, et al. Systems perspectives on erythromycin biosynthesis by comparative genomic and transcriptomic analyses of S. erythraea E3 and NRRL23338 strains[J]. BMC Genomics, 2013, 14: 523. |
| 26 | Zhang Y P, Liu X M, Yin T, et al. Comparative transcriptomic analysis of two Saccharopolysporaspinosa strains reveals the relationships between primary metabolism and spinosad production[J]. Scientific Reports, 2021, 11: 14779. |
| 27 | Zhao F L, Xue C Y, Wang M L, et al. A comparative metabolomics analysis of Saccharopolyspora spinosa WT, WH124, and LU104 revealed metabolic mechanisms correlated with increases in spinosad yield[J]. Bioscience, Biotechnology, and Biochemistry, 2013, 77(8): 1661-1668. |
| 28 | Luo Y S, Ding X Z, Xia L Q, et al. Comparative proteomic analysis of Saccharopolyspora spinosa SP06081 and PR2 strains reveals the differentially expressed proteins correlated with the increase of spinosad yield[J]. Proteome Science, 2011, 9: 40. |
| 29 | 陈爽, 赵晨, 黎琪, 等. 丁烯基多杀菌素高产菌株的诱变选育及培养基优化[J]. 江苏农业科学, 2018, 46(9): 108-111. |
| Chen S, Zhao C, Li Q, et al. Mutation breeding and medium optimization of butenyl-spinosyns producing strain[J]. Jiangsu Agricultural Sciences, 2018, 46(9): 108-111. | |
| 30 | 郭超, 王超, 郭伟群. 一种多杀菌素糖苷配基半抗原及其制备方法和应用: 112390776A[P]. 2021-02-23. |
| Guo C, Wang C, Guo W Q. Spinosad aglycone hapten, preparation method and application thereof: 112390776A[P]. 2021-02-23. | |
| 31 | Lan J Q, Zhao H W, Jin X T, et al. Development of a monoclonal antibody-based immunoaffinity chromatography and a sensitive immunoassay for detection of spinosyn A in milk, fruits, and vegetables[J]. Food Control, 2019, 95: 196-205. |
| 32 | 罗林根, 杨燕, 魏慧, 等. 须糖多孢菌Saccharopolyspora pogona的核糖体工程改造对丁烯基多杀菌素合成的影响[J]. 生物工程学报, 2016, 32(2): 259-263. |
| Luo L G, Yang Y, Wei H, et al. Effect of ribosome engineering on butenyl-spinosyns synthesis of Saccharopolyspora pogona[J]. Chinese Journal of Biotechnology, 2016, 32(2): 259-263. | |
| 33 | Zhang Q, Ren J W, Wang W S, et al. A versatile transcription-translation in one approach for activation of cryptic biosynthetic gene clusters [J]. ACS Chemical Biology, 2020, 15(9): 2551-2557. |
| 34 | 邬洋, 徐妙, 罗林根, 等. 丁烯基多杀菌素高产菌株的巴龙霉素抗性筛选[J]. 中国生物防治学报, 2015, 31(1): 106-114. |
| Wu Y, Xu M, Luo L G, et al. Screening of butenyl-spinosyn high-yield strains by paromomycin resistance[J]. Chinese Journal of Biological Control, 2015, 31(1): 106-114. | |
| 35 | Guo C, Guo W Q, Liu Y C, et al. Complete genome sequence of butenyl-spinosyn-producing Saccharopolyspora strain ASAGF58[J]. Annals of Microbiology, 2020, 70(1): 1-6. |
| 36 | Huang K X, Xia L Q, Zhang Y M, et al. Recent advances in the biochemistry of spinosyns[J]. Applied Microbiology and Biotechnology, 2009, 82(1): 13-23. |
| 37 | 赵方龙. 刺糖多孢菌胞内代谢物LC-MS分析与多杀菌素分离纯化工艺研究[D]. 天津: 天津大学, 2013. |
| Zhao F L. Study of intracellular metabolites of Saccharopolysporaspinosa using LC-MS and separation process of spinosyn[D]. Tianjin: Tianjin University, 2013. | |
| 38 | 薛程斌, 赵春田, 裘娟萍. 提高多杀菌素合成前体: 乙酰辅酶A流量的代谢调控策略[J]. 科技通报, 2018, 34(7): 9-15. |
| Xue C B, Zhao C T, Qiu J P. Metabolic control strategy of increasing spinosad biosynthesis precursor-acetyl CoA flow[J]. Bulletin of Science and Technology, 2018, 34(7): 9-15. | |
| 39 | Liu Z D, Zhu Z R, Tang J L, et al. RNA-seq-based transcriptomic analysis of Saccharopolyspora spinosa revealed the critical function of PEP phosphonomutase in the replenishment pathway[J]. Journal of Agricultural and Food Chemistry, 2020, 68(49): 14660-14669. |
| 40 | Liu Z D, Xiao J, Tang J L, et al. Effects of acuC on the growth development and spinosad biosynthesis of Saccharopolyspora spinosa[J]. Microbial Cell Factories, 2021, 20(1):141. |
| 41 | Huang Y, Zhang X L, Zhao C, et al. Improvement of spinosad production upon utilization of oils and manipulation of β-oxidation in a high-producing Saccharopolyspora spinosa strain[J]. Journal of Molecular Microbiology and Biotechnology, 2018, 28(2): 53-64. |
| 42 | 徐妙, 邬洋, 杨燕, 等. 环腺苷酸受体蛋白基因的过表达对刺糖多孢菌生长和多杀菌素合成的影响[J]. 中国农业科学, 2014, 47(18): 3577-3587. |
| Xu M, Wu Y, Yang Y, et al. Impact on strain growth and spinosad biosynthesis by overexpression of cyclic AMP receptor protein gene in Saccharopolyspora spinosa[J]. Scientia Agricultura Sinica, 2014, 47(18): 3577-3587. | |
| 43 | 杨燕, 罗林根, 徐妙, 等. 亮氨酰氨肽酶基因的阻断对刺糖多孢菌生长及次级代谢产物合成的影响[J]. 微生物学报, 2016, 56(4): 629-642. |
| Yang Y, Luo L G, Xu M, et al. Disruption of leucyl aminopeptidase gene affects phenotypes and second metabolite production of Saccharopolyspora spinosa[J]. Acta Microbiologica Sinica, 2016, 56(4): 629-642. | |
| 44 | 刘红雪, 蔡妹, 张由恒, 等. 磷酸甘露酶基因的阻断对刺糖多孢菌的形态及其多杀菌素合成的影响[J]. 中国生物防治学报, 2017, 33(1): 134-141. |
| Liu H X, Cai M, Zhang Y H, et al. Impact on strain phenotypes and spinosad biosynthesis by disruption of phosphomannomutase gene in Saccharopolyspora spinosa[J]. Chinese Journal of Biological Control, 2017, 33(1): 134-141. | |
| 45 | 肖洁, 刘朱东, 彭胜男, 等. glnA基因对刺糖多孢菌生长发育及多杀菌素合成的影响[J]. 中国生物防治学报, 2018, 34(4): 625-638. |
| Xiao J, Liu Z D, Peng S N, et al. The effect of glnA gene on growth development and spinosad biosynthesis in Saccharopolyspora spinosa[J]. Chinese Journal of Biological Control, 2018, 34(4): 625-638. | |
| 46 | 彭胜男, 何昊城, 苑爽芹, 等. fcl基因对须糖多孢菌丁烯基多杀菌素生物合成及生长发育的影响[J]. 生物工程学报, 2019, 35(9): 1662-1675. |
| Peng S N, He H C, Yuan S Q, et al. Effect of fcl gene for butenyl-spinosyn biosynthesis and growth of Saccharopolyspora pogona[J]. Chinese Journal of Biotechnology, 2019, 35(9): 1662-1675. | |
| 47 | Tang J L, Zhu Z R, He H C, et al. Bacterioferritin: a key iron storage modulator that affects strain growth and butenyl-spinosyn biosynthesis in Saccharopolyspora pogona[J]. Microbial Cell Factories, 2021, 20(1): 157. |
| 48 | Rang J, Li Y L, Cao L, et al. Deletion of a hybrid NRPS-T1PKS biosynthetic gene cluster via Latour gene knockout system in Saccharopolyspora pogona and its effect on butenyl-spinosyn biosynthesis and growth development[J]. Microbial Biotechnology, 2021, 14(6): 2369-2384. |
| 49 | Wang X Y, Zhang C B, Wang M L, et al. Genome-scale metabolic network reconstruction of Saccharopolyspora spinosa for spinosad production improvement[J]. Microbial Cell Factories, 2014, 13(1): 41. |
| 50 | 黄颖, 赵晨, 杨博磊, 等. 刺糖多孢菌高产菌株和野生型菌株多杀菌素生物合成基因簇(spn)在发酵过程中的表达分析[J]. 农业生物技术学报, 2014, 22(11): 1337-1346. |
| Huang Y, Zhao C, Yang B L, et al. Expression analysis of spinosad biosynthetic gene cluster(spn) in hyperproducing and wild-type Saccharopolyspora spinosa strains during fermentation[J]. Journal of Agricultural Biotechnology, 2014, 22(11): 1337-1346. | |
| 51 | Xue C Y, Duan Y J, Zhao F L, et al. Stepwise increase of spinosad production in Saccharopolyspora spinosa by metabolic engineering[J]. Biochemical Engineering Journal, 2013, 72: 90-95. |
| 52 | Tang Y, Xia L Q, Ding X Z, et al. Duplication of partial spinosyn biosynthetic gene cluster in Saccharopolyspora spinosa enhances spinosyn production[J]. FEMS Microbiology Letters, 2011, 325(1): 22-29. |
| 53 | Pan H X, Li J A, He N J, et al. Improvement of spinosad production by overexpression of gtt and gdh controlled by promoter PermE* in Saccharopolyspora spinosa SIPI-A2090[J]. Biotechnology Letters, 2011, 33(4): 733-739. |
| 54 | Rang J, He H C, Yuan S Q, et al. Deciphering the metabolic pathway difference between Saccharopolyspora pogona and Saccharopolyspora spinosa by comparative proteomics and metabonomics[J]. Frontiers in Microbiology, 2020, 11: 396. |
| 55 | Li L, Rang J, He H C, et al. Impact on strain growth and butenyl-spinosyn biosynthesis by overexpression of polynucleotide phosphorylase gene in Saccharopolyspora pogona[J]. Applied Microbiology and Biotechnology, 2018, 102(18): 8011-8021. |
| 56 | Li L, Gong L, He H C, et al. AfsR is an important regulatory factor for growth and butenyl-spinosyn biosynthesis of Saccharopolyspora pogona[J]. Annals of Microbiology, 2019, 69(8): 809-818. |
| 57 | 何思颖, 柏丹, 夏伦, 等. PadR对须糖多孢菌丁烯基多杀菌素生物合成及转运相关蛋白表达的影响[J]. 湖南师范大学自然科学学报, 2019, 42(5): 44-51. |
| He S Y, Bai D, Xia L, et al. The impact of PadR on butenyl-spinosyn biosynthesis and transporters expression in Saccharopolyspora pogona[J]. Journal of Natural Science of Hunan Normal University, 2019, 42(5): 44-51. | |
| 58 | He H C, Yuan S Q, Hu J J, et al. Effect of the TetR family transcriptional regulator Sp1418 on the global metabolic network of Saccharopolyspora pogona[J]. Microbial Cell Factories, 2020, 19(1): 27. |
| 59 | Rang J, He H C, Chen J M, et al. SenX3-RegX3, an important two-component system, regulates strain growth and butenyl-spinosyn biosynthesis in Saccharopolyspora pogona[J]. iScience, 2020, 23(8): 101398. |
| 60 | He H C, Peng S N, Yuan S Q, et al. Effects of lytS-L on the primary metabolism and butenyl-spinosyn biosynthesis in Saccharopolyspora pogona[J]. Gene, 2021, 766: 145130. |
| 61 | Rang J, Zhu Z R, Li Y L, et al. Identification of a TetR family regulator and a polyketide synthase gene cluster involved in growth development and butenyl-spinosyn biosynthesis of Saccharopolyspora pogona[J]. Applied Microbiology and Biotechnology, 2021, 105(4): 1519-1533. |
| 62 | Liu R, Deng Z X, Liu T G. Streptomyces species: ideal chassis for natural product discovery and overproduction[J]. Metabolic Engineering, 2018, 50: 74-84. |
| 63 | Zhang J, Zhang D, Zhu J, et al. Efficient multiplex genome editing in streptomyces via engineered CRISPR-Cas12a systems[J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 726. |
| 64 | Song C Y, Luan J, Li R J, et al. RedEx: a method for seamless DNA insertion and deletion in large multimodular polyketide synthase gene clusters[J]. Nucleic Acids Research, 2020, 48(22): e130. |
| 65 | Tan G Y, Deng K H, Liu X H, et al. Heterologous biosynthesis of spinosad: an omics-guided large, polyketide synthase gene cluster reconstitution in Streptomyces[J]. ACS Synthetic Biology, 2017, 6(6): 995-1005. |
| 66 | Zhao C, Huang Y, Guo C, et al. Heterologous expression of spinosyn biosynthetic gene cluster in Streptomyces species is dependent on the expression of rhamnose biosynthesis genes[J]. Journal of Molecular Microbiology and Biotechnology, 2017, 27(3): 190-198. |
| 67 | Song C Y, Luan J, Cui Q W, et al. Enhanced heterologous spinosad production from a 79-kb synthetic multioperon assembly[J]. ACS Synthetic Biology, 2019, 8(1): 137-147. |
| 68 | Wang H L, Li Z, Jia R N, et al. ExoCET: exonuclease in vitro assembly combined with RecET recombination for highly efficient direct DNA cloning from complex genomes[J]. Nucleic Acids Research, 2018, 46(5): e28. |
| [1] | 刘昕, 戈钧, 李春. 光驱动微生物杂合系统提高生物制造水平[J]. 化工学报, 2023, 74(1): 330-341. |
| [2] | 刘雪, 张莉娟, 赵广荣. 大肠杆菌偏利共培养系统合成大豆苷元[J]. 化工学报, 2022, 73(9): 4015-4024. |
| [3] | 孙怡, 张腾, 吕波, 李春. 胞内生物传感器提高微生物细胞工厂的精细调控[J]. 化工学报, 2022, 73(2): 521-534. |
| [4] | 王欣慧, 王颖, 姚明东, 肖文海. 维生素A生物合成的研究进展[J]. 化工学报, 2022, 73(10): 4311-4323. |
| [5] | 周武林, 高惠芳, 吴玉玲, 张显, 徐美娟, 杨套伟, 邵明龙, 饶志明. 重组酿酒酵母生物合成菜油甾醇[J]. 化工学报, 2021, 72(8): 4314-4324. |
| [6] | 杨瑞雄, 郑鑫, 陆涛, 赵誉泽, 杨庆华, 卢英华, 何宁, 凌雪萍. 烯酰还原酶基因的替换对裂殖壶菌合成二十碳五烯酸的影响[J]. 化工学报, 2021, 72(7): 3768-3779. |
| [7] | 陈婷婷, 韩恺忻, 陈翠雪, 凌雪萍, 沈亮, 卢英华. 铁还原菌Shewanella xiamenensis BC01的有机溶剂应激研究[J]. 化工学报, 2021, 72(7): 3747-3756. |
| [8] | 毛金竹, 肖淑玲, 杨智淳, 王孝宇, 张诗, 陈俊宏, 谢佶晟, 陈福德, 黄子诺, 冯天宇, 张瑷珲, 方柏山. 合成生物学在农残检测领域的应用[J]. 化工学报, 2021, 72(5): 2413-2425. |
| [9] | 王欣, 赵鹏, 李清扬, 田平芳. 半导体合成生物学的研究进展[J]. 化工学报, 2021, 72(5): 2426-2435. |
| [10] | 苏楠, 吴亦楠, 陈韵亿, 金丽华, 张翀, Aikawa Shimpei, Hasunuma Tomohisa, Kondo Akihiko, 邢新会. ARTP诱变钝顶螺旋藻突变体比较组学研究[J]. 化工学报, 2021, 72(12): 6298-6310. |
| [11] | 赵贞尧, 张保财, 李锋, 宋浩. 产电细胞的合成生物学设计构建[J]. 化工学报, 2021, 72(1): 468-482. |
| [12] | 王炼, 吴迪, 周景文. 木脂素的生物合成及其微生物法生产的研究进展[J]. 化工学报, 2021, 72(1): 320-333. |
| [13] | 江龙, 王开杰, 孔晴, 陆晟, 陈小强. 基于金刚烷-二氧杂环丁烷化学发光探针的研究进展[J]. 化工学报, 2021, 72(1): 229-246. |
| [14] | 王凯峰, 王金鹏, 韦萍, 纪晓俊. 代谢工程改造解脂耶氏酵母生产脂肪酸及其衍生物[J]. 化工学报, 2021, 72(1): 351-365. |
| [15] | 秦磊, 俞杰, 宁小钰, 孙文涛, 李春. 合成生物系统构建与绿色生物“智”造[J]. 化工学报, 2020, 71(9): 3979-3994. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备 11010102001995号
京公网安备 11010102001995号