化工学报 ›› 2021, Vol. 72 ›› Issue (12): 6298-6310.DOI: 10.11949/0438-1157.20211340
苏楠1( ),吴亦楠1(
),吴亦楠1( ),陈韵亿1,金丽华2,张翀1,Aikawa Shimpei3,Hasunuma Tomohisa3,Kondo Akihiko3,邢新会1,4,5(
),陈韵亿1,金丽华2,张翀1,Aikawa Shimpei3,Hasunuma Tomohisa3,Kondo Akihiko3,邢新会1,4,5( )
)
收稿日期:2021-09-16
修回日期:2021-11-23
出版日期:2021-12-05
发布日期:2021-12-22
通讯作者:
邢新会
作者简介:苏楠(1983—),男,博士,基金资助:
Nan SU1( ),Yinan WU1(
),Yinan WU1( ),Yinyee TAN1,Lihua JIN2,Chong ZHANG1,Aikawa SHIMPEI3,Hasunuma TOMOHISA3,Kondo AKIHIKO3,Xinhui XING1,4,5(
),Yinyee TAN1,Lihua JIN2,Chong ZHANG1,Aikawa SHIMPEI3,Hasunuma TOMOHISA3,Kondo AKIHIKO3,Xinhui XING1,4,5( )
)
Received:2021-09-16
Revised:2021-11-23
Online:2021-12-05
Published:2021-12-22
Contact:
Xinhui XING
摘要:
微藻作为地球上重要的生物资源,为水圈提供了大量的初级代谢产物,是合成生物学和生物制造研究和应用的重要底盘微生物。其中,钝顶螺旋藻(Spirulina platensis)具有多糖含量高、营养价值高、培养工艺成熟、应用范围广等特点,其诱变育种及比较组学研究可为微藻细胞工厂系统改造技术发展提供重要依据。本课题组前期通过常压室温等离子体(atmospheric and room temperature plasmas,ARTP)诱变方法获得了三株钝顶螺旋藻突变体。本研究在连续光照培养条件下,对三株突变体的重要生理特征进行了表征,发现突变株具有高絮凝表型,且重要化合物含量也与野生型具有一定的差异。进一步,本研究通过对其主要代谢产物的代谢组学分析和全基因组测序,对突变表型产生的机理进行了初步解析。
中图分类号:
苏楠, 吴亦楠, 陈韵亿, 金丽华, 张翀, Aikawa Shimpei, Hasunuma Tomohisa, Kondo Akihiko, 邢新会. ARTP诱变钝顶螺旋藻突变体比较组学研究[J]. 化工学报, 2021, 72(12): 6298-6310.
Nan SU, Yinan WU, Yinyee TAN, Lihua JIN, Chong ZHANG, Aikawa SHIMPEI, Hasunuma TOMOHISA, Kondo AKIHIKO, Xinhui XING. Comparative omics study of Spirulinaplatensis mutants based on ARTP mutagenesis breeding system[J]. CIESC Journal, 2021, 72(12): 6298-6310.
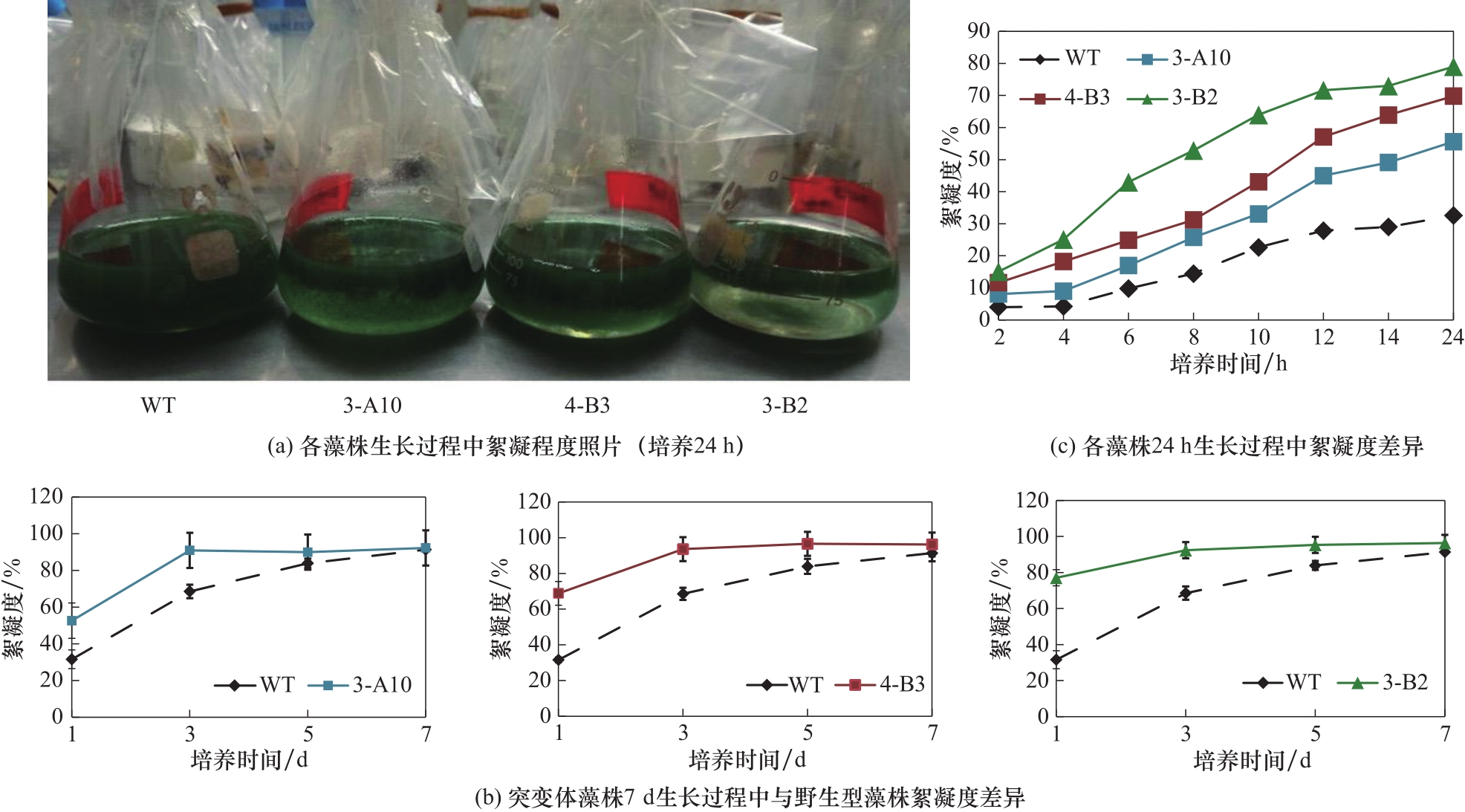
图2 突变体藻株在生长过程中的絮凝度变化: (a) 各藻株摇床培养24 h后,静置5 min时的照片; (b) 各藻株摇床7 d培养过程中的絮凝度变化; (c) 经24 h摇床培养的藻株,静置培养不同时间后的絮凝度
Fig.2 Change of flocculation over culture time. (a) Pictures of S. platensis strain cultures after 24 h incubation in the shaker; (b) The change of flocculation of strain cultures during 7 d of incubation in the shaker; (c) The change of flocculation of strain cultures (harvested after 24 h of incubation in the shaker) after stationary sedimentation in tubes for different time periods

图7 测定中间代谢产物在各代谢途径中的分布红色标记中间产物为突变体与野生型含量有较显著差异的代谢产物;黄色标记中间产物为突变体与野生型含量没有显著差异的代谢产物Ac-CoA —乙酰辅酶A(acetyl-CoA); ADP-GLC —ADP-葡萄糖(ADP-glucose); CA —柠檬酸盐(citrate); E4P —赤藻糖(erythrose-4-phosphate); F6P —果糖-6-磷酸(fructose-6-phosphate); FUM —延胡索酸(fumarate); GAP —甘油醛-3-磷酸(glyceraldehyde-3-phosphate); G1P —葡萄糖-1-磷酸(glucose-1-phosphate); G3P —甘油-3-磷酸(glycerol-3-phosphate); G6P —葡萄糖-6-磷酸(glucose-6-phosphate); ICTT —异柠檬酸(isocitrate); LPA —溶血磷脂酸(lysobisphosphatidic acids); α-KG —α-酮戊二酸(2-ketoglutarate); MAA —苹果酸盐(malate); OAA —草酰乙酸盐(oxaloacetate); PEP —磷酸烯醇丙酮酸盐(phosphoenolpyruvate); PYR —丙酮酸(pyruvic acid); 3PGA —3-磷酸甘油酸(3-phosphoglycerate); R5P —核糖-5-磷酸(ribose-5-phosphate); Ru5P —核酮糖-5-磷酸(ribulose-5-phosphate); RuBP —核酮糖-1,5-二磷酸(ribulose-1,5-bisphosphate); S7P —景天庚糖-7-磷酸(sedoheptulose-7-phosphate); SBP —景天庚糖-1,7-二磷酸(sedoheptulose-1,7-bisphosphate); SSA —琥珀酸辅酶A(succinate-CoA); SUCC —琥珀酸(succinate); UDP-GLC—UDP-葡萄糖(UDP-glucose); X5P —木酮糖-5-磷酸(xylulose-5-phosphate)
Fig.7 Location of detected important metabolites in strain metabolic networks
| 聚类分析 | 3-A10 | 3-B2 | 4-B3 |
|---|---|---|---|
| 生物学过程相关 (BP) | 910 | 913 | 910 |
| 细胞组分相关 (CC) | 103 | 101 | 101 |
| 分子功能相关 (MF) | 668 | 672 | 667 |
表1 突变体突变基因GO聚类分析
Table 1 Gene Ontology clustering analysis of mutated strains
| 聚类分析 | 3-A10 | 3-B2 | 4-B3 |
|---|---|---|---|
| 生物学过程相关 (BP) | 910 | 913 | 910 |
| 细胞组分相关 (CC) | 103 | 101 | 101 |
| 分子功能相关 (MF) | 668 | 672 | 667 |
| 序列差异性 | 对照 | 3-A10 | 3-B2 | 4-B3 |
|---|---|---|---|---|
| 单碱基突变数量 | 241873 | 243818 | 241020 | 244320 |
| 过滤野生型基因后的数量 | — | 4063 | 2532 | 4536 |
| 同义单碱基突变 | — | 1040 | 681 | 1229 |
| 错义单碱基突变 | — | 1591 | 1059 | 1823 |
| 进化率 | — | 1.53 | 1.55 | 1.48 |
| 序列插入突变数量 | 345 | 353 | 330 | 362 |
| 过滤野生型基因后的数量 | — | 134 | 101 | 123 |
| 错义插入突变 | — | 82 | 56 | 81 |
表A1 螺旋藻各藻株基因组测序突变数量统计
Table A1 Statistics on the number of mutations in genome sequencing of Spirulina platensis
| 序列差异性 | 对照 | 3-A10 | 3-B2 | 4-B3 |
|---|---|---|---|---|
| 单碱基突变数量 | 241873 | 243818 | 241020 | 244320 |
| 过滤野生型基因后的数量 | — | 4063 | 2532 | 4536 |
| 同义单碱基突变 | — | 1040 | 681 | 1229 |
| 错义单碱基突变 | — | 1591 | 1059 | 1823 |
| 进化率 | — | 1.53 | 1.55 | 1.48 |
| 序列插入突变数量 | 345 | 353 | 330 | 362 |
| 过滤野生型基因后的数量 | — | 134 | 101 | 123 |
| 错义插入突变 | — | 82 | 56 | 81 |
| 代谢途径 | 突变体 | 代谢途径 | 突变体 | 代谢途径 | 突变体 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 3-A10 | 3-B2 | 4-B3 | 3-A10 | 3-B2 | 4-B3 | 3-A10 | 3-B2 | 4-B3 | |||
| 糖酵解 | 9 | 9 | 9 | 苯丙氨酸代谢 | 2 | 2 | 2 | 乙醛酸、二羧酸代谢 | 4 | 4 | 4 |
| 三羧酸循环 | 2 | 2 | 2 | 色氨酸代谢 | 1 | 1 | 1 | 丙酸代谢 | 1 | 1 | 1 |
| 磷酸戊糖途径 | 7 | 6 | 7 | 苯丙氨酸、酪氨酸、色氨酸合成 | 10 | 10 | 10 | 苯乙烯降解 | 1 | 1 | 1 |
| 果糖、甘露糖代谢 | 5 | 5 | 5 | 叶酸合成 | 8 | 8 | 8 | ||||
| 半乳糖代谢 | 4 | 4 | 4 | 新生毒素生物合成 | 1 | 1 | 1 | 甲烷代谢 | 6 | 6 | 6 |
| 脂肪酸合成 | 3 | 3 | 3 | β-丙氨酸代谢 | 2 | 2 | 2 | 碳固定及光合作用 | 8 | 8 | 8 |
| 氧化磷酸化 | 5 | 5 | 5 | 牛磺酸代谢 | 1 | 1 | 1 | 原核生物碳固定途径 | 6 | 5 | 5 |
| 光合作用 | 2 | 2 | 2 | 有机含硒化合物代谢 | 2 | 2 | 2 | 硫胺素代谢 | 3 | 3 | 3 |
| 嘌呤代谢 | 17 | 17 | 17 | 氨基甲酸代谢 | 2 | 2 | 2 | 核黄素代谢 | 7 | 7 | 7 |
| 丙氨酸、天冬氨酸、谷氨酸代谢 | 5 | 5 | 4 | D-谷氨酰胺、D-谷氨酸代谢 | 1 | 1 | 1 | 维生素B6代谢 | 1 | 1 | 1 |
| 烟酸盐、烟酰胺代谢 | 3 | 3 | 3 | ||||||||
| 四环素合成 | 1 | 1 | 1 | D-丙氨酸代谢 | 1 | 1 | 1 | 泛酸盐及辅酶a生物合成 | 8 | 8 | 8 |
| 黄曲霉毒素合成 | 1 | 1 | 1 | 谷胱甘肽代谢 | 7 | 7 | 7 | ||||
| 甘氨酸、丝氨酸、苏氨酸代谢 | 4 | 4 | 4 | 淀粉、蔗糖代谢 | 7 | 7 | 7 | 生物素代谢 | 1 | 1 | 1 |
| 氨基糖、核苷酸糖代谢 | 6 | 6 | 6 | 叶酸生物合成 | 3 | 3 | 3 | ||||
| 半胱氨酸、蛋氨酸代谢 | 6 | 5 | 6 | 链霉素生物合成 | 5 | 5 | 5 | 阿特拉津降解 | 1 | 1 | 1 |
| 缬氨酸、亮氨酸、异亮氨酸生物合成 | 2 | 2 | 2 | 聚酮糖单元生物合成 | 2 | 2 | 2 | 卟啉、叶绿素代谢 | 15 | 15 | 15 |
| 丁松香和新霉素生物合成 | 2 | 2 | 2 | 萜类化合物生物合成 | 3 | 3 | 3 | ||||
| 赖氨酸生物合成 | 2 | 2 | 2 | ||||||||
| 精氨酸、脯氨酸代谢 | 8 | 8 | 8 | 脂多糖生物合成 | 1 | 1 | 0 | 氮代谢 | 7 | 7 | 7 |
| 组氨酸代谢 | 4 | 4 | 4 | 肽聚糖生物合成 | 3 | 3 | 3 | 糖代谢 | 6 | 6 | 6 |
| 氨基苯甲酸降解 | 2 | 2 | 甘油酯代谢 | 2 | 2 | 2 | 苯丙素生物合成 | 1 | 1 | 1 | |
| 药物代谢相关酶类 | 2 | 2 | 2 | 磷酸肌醇代谢 | 2 | 2 | 2 | 氨酰基-tRNA生物合成 | 6 | 6 | 6 |
| 磷脂酰肌醇信号系统 | 1 | 1 | 1 | 甘油磷脂代谢 | 4 | 4 | 4 | ||||
| 丙酮酸代谢 | 7 | 7 | 7 | 鞘脂类代谢 | 2 | 2 | 2 | 细胞色素P450代谢 | 1 | 1 | 1 |
| 鞘糖脂生物合成 | 1 | 1 | 1 | 鞘糖脂生物合成乳糖 | 1 | 1 | 1 | ||||
表A2 突变体突变基因所属代谢网络分析
Table A2 Metabolic network analysis of mutant genes
| 代谢途径 | 突变体 | 代谢途径 | 突变体 | 代谢途径 | 突变体 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 3-A10 | 3-B2 | 4-B3 | 3-A10 | 3-B2 | 4-B3 | 3-A10 | 3-B2 | 4-B3 | |||
| 糖酵解 | 9 | 9 | 9 | 苯丙氨酸代谢 | 2 | 2 | 2 | 乙醛酸、二羧酸代谢 | 4 | 4 | 4 |
| 三羧酸循环 | 2 | 2 | 2 | 色氨酸代谢 | 1 | 1 | 1 | 丙酸代谢 | 1 | 1 | 1 |
| 磷酸戊糖途径 | 7 | 6 | 7 | 苯丙氨酸、酪氨酸、色氨酸合成 | 10 | 10 | 10 | 苯乙烯降解 | 1 | 1 | 1 |
| 果糖、甘露糖代谢 | 5 | 5 | 5 | 叶酸合成 | 8 | 8 | 8 | ||||
| 半乳糖代谢 | 4 | 4 | 4 | 新生毒素生物合成 | 1 | 1 | 1 | 甲烷代谢 | 6 | 6 | 6 |
| 脂肪酸合成 | 3 | 3 | 3 | β-丙氨酸代谢 | 2 | 2 | 2 | 碳固定及光合作用 | 8 | 8 | 8 |
| 氧化磷酸化 | 5 | 5 | 5 | 牛磺酸代谢 | 1 | 1 | 1 | 原核生物碳固定途径 | 6 | 5 | 5 |
| 光合作用 | 2 | 2 | 2 | 有机含硒化合物代谢 | 2 | 2 | 2 | 硫胺素代谢 | 3 | 3 | 3 |
| 嘌呤代谢 | 17 | 17 | 17 | 氨基甲酸代谢 | 2 | 2 | 2 | 核黄素代谢 | 7 | 7 | 7 |
| 丙氨酸、天冬氨酸、谷氨酸代谢 | 5 | 5 | 4 | D-谷氨酰胺、D-谷氨酸代谢 | 1 | 1 | 1 | 维生素B6代谢 | 1 | 1 | 1 |
| 烟酸盐、烟酰胺代谢 | 3 | 3 | 3 | ||||||||
| 四环素合成 | 1 | 1 | 1 | D-丙氨酸代谢 | 1 | 1 | 1 | 泛酸盐及辅酶a生物合成 | 8 | 8 | 8 |
| 黄曲霉毒素合成 | 1 | 1 | 1 | 谷胱甘肽代谢 | 7 | 7 | 7 | ||||
| 甘氨酸、丝氨酸、苏氨酸代谢 | 4 | 4 | 4 | 淀粉、蔗糖代谢 | 7 | 7 | 7 | 生物素代谢 | 1 | 1 | 1 |
| 氨基糖、核苷酸糖代谢 | 6 | 6 | 6 | 叶酸生物合成 | 3 | 3 | 3 | ||||
| 半胱氨酸、蛋氨酸代谢 | 6 | 5 | 6 | 链霉素生物合成 | 5 | 5 | 5 | 阿特拉津降解 | 1 | 1 | 1 |
| 缬氨酸、亮氨酸、异亮氨酸生物合成 | 2 | 2 | 2 | 聚酮糖单元生物合成 | 2 | 2 | 2 | 卟啉、叶绿素代谢 | 15 | 15 | 15 |
| 丁松香和新霉素生物合成 | 2 | 2 | 2 | 萜类化合物生物合成 | 3 | 3 | 3 | ||||
| 赖氨酸生物合成 | 2 | 2 | 2 | ||||||||
| 精氨酸、脯氨酸代谢 | 8 | 8 | 8 | 脂多糖生物合成 | 1 | 1 | 0 | 氮代谢 | 7 | 7 | 7 |
| 组氨酸代谢 | 4 | 4 | 4 | 肽聚糖生物合成 | 3 | 3 | 3 | 糖代谢 | 6 | 6 | 6 |
| 氨基苯甲酸降解 | 2 | 2 | 甘油酯代谢 | 2 | 2 | 2 | 苯丙素生物合成 | 1 | 1 | 1 | |
| 药物代谢相关酶类 | 2 | 2 | 2 | 磷酸肌醇代谢 | 2 | 2 | 2 | 氨酰基-tRNA生物合成 | 6 | 6 | 6 |
| 磷脂酰肌醇信号系统 | 1 | 1 | 1 | 甘油磷脂代谢 | 4 | 4 | 4 | ||||
| 丙酮酸代谢 | 7 | 7 | 7 | 鞘脂类代谢 | 2 | 2 | 2 | 细胞色素P450代谢 | 1 | 1 | 1 |
| 鞘糖脂生物合成 | 1 | 1 | 1 | 鞘糖脂生物合成乳糖 | 1 | 1 | 1 | ||||
| 1 | Miyamoto K. Renewable Biological Systems for Alternative Sustainable Energy Production[M]. Netherlands: Elsevier Ltd., 1997. |
| 2 | Jagadevan S, Banerjee A, Banerjee C, et al. Recent developments in synthetic biology and metabolic engineering in microalgae towards biofuel production[J]. Biotechnology for Biofuels, 2018, 11: 185. |
| 3 | Casazza A A, Spennati E, Converti A, et al. Production of carbon-based biofuels by pyrolysis of exhausted Arthrospira platensis biomass after protein or lipid recovery[J]. Fuel Processing Technology, 2020, 201: 106336. |
| 4 | Khan M I, Shin J H, Kim J D. The promising future of microalgae: current status, challenges, and optimization of a sustainable and renewable industry for biofuels, feed, and other products[J]. Microbial Cell Factories, 2018, 17(1): 36. |
| 5 | Liestianty D, Rodianawati I, Arfah R A, et al. Nutritional analysis of Spirulina sp. to promote as superfood candidate[J]. IOP Conference Series: Materials Science and Engineering, 2019, 509: 012031. |
| 6 | Nasirian F, Dadkhah M, Moradi-Kor N, et al. Effects of Spirulina platensis microalgae on antioxidant and anti-inflammatory factors in diabetic rats[J]. Diabetes, Metabolic Syndrome and Obesity: Targets and Therapy, 2018, 11: 375-380. |
| 7 | Aydi S S, Aydi S, Ben Abdallah Kolsi R, et al. CO2 enrichment: enhancing antioxidant, antibacterial and anticancer activities in Arthrospira platensis[J]. Food Bioscience, 2020, 35: 100575. |
| 8 | Saranraj P, Sivasakthi S. Spirulina platensis-food for future: a review[J]. Asian J. Pharm. Sci. Technol., 2014, 4(1): 26-33. |
| 9 | Mukhopadhyay C D. Engineering spirulina for enhanced medicinal application[M]//Algal Biorefinery: An Integrated Approach. Cham: Springer International Publishing, 2015: 235-252. |
| 10 | Saini D K, Chakdar H, Pabbi S, et al. Enhancing production of microalgal biopigments through metabolic and genetic engineering[J]. Critical Reviews in Food Science and Nutrition, 2020, 60(3): 391-405. |
| 11 | Ragusa I, Nardone G N, Zanatta S, et al. Spirulina for skin care: a bright blue future[J]. Cosmetics, 2021, 8(1): 7. |
| 12 | Rempel A, de Souza Sossella F, Margarites A C, et al. Bioethanol from Spirulina platensis biomass and the use of residuals to produce biomethane: an energy efficient approach[J]. Bioresource Technology, 2019, 288: 121588. |
| 13 | Kawata Y, Yano S, Kojima H, et al. Transformation of Spirulina platensis strain C1 (Arthrospira sp. PCC9438) with Tn5 transposase-transposon DNA-cation liposome complex[J]. Marine Biotechnology, 2004, 6(4): 355-363. |
| 14 | Dehghani J, Adibkia K, Movafeghi A, et al. Stable transformation of Spirulina (Arthrospira) platensis: a promising microalga for production of edible vaccines[J]. Applied Microbiology and Biotechnology, 2018, 102(21): 9267-9278. |
| 15 | Jeamton W, Dulsawat S, Tanticharoen M, et al. Overcoming intrinsic restriction enzyme barriers enhances transformation efficiency in Arthrospira platensis C1[J]. Plant and Cell Physiology, 2017, 58(4): 822-830. |
| 16 | Cheevadhanarak S, Paithoonrangsarid K, Prommeenate P, et al. Draft genome sequence of Arthrospira platensis C1 (PCC9438)[J]. Standards in Genomic Sciences, 2012, 6(1): 43-53. |
| 17 | 殷春涛, 胡鸿钧, 李夜光, 等. 中温螺旋藻新品系的选育研究[J]. 武汉植物学研究, 1997, 15(3): 250-254. |
| Yin C T, Hu H J, Li Y G, et al. Studies on middle temperature strains selection of Spirulina platensis[J]. Journal of Wuhan Botanical Research, 1997, 15(3): 250-254. | |
| 18 | 李建宏, 郑卫, 倪霞, 等. 两株钝顶螺旋藻紫外诱变株的特征[J]. 水生生物学报, 2001, 25(5): 486-490. |
| Li J H, Zheng W, Ni X, et al. Characteristics of two Spirulina platensis mutants induced by ultraviolet[J]. Acta Hydrobiologica Sinica, 2001, 25(5): 486-490. | |
| 19 | Fang M, Jin L, Zhang C, et al. Rapid mutation of Spirulina platensis by a new mutagenesis system of atmospheric and room temperature plasmas (ARTP) and generation of a mutant library with diverse phenotypes[J]. PLoS One, 2013, 8(10): e77046. |
| 20 | Shirnalli G G, Kaushik M S, Kumar A, et al. Isolation and characterization of high protein and phycocyanin producing mutants of Arthrospira platensis[J]. Journal of Basic Microbiology, 2018, 58(2): 162-171. |
| 21 | Cheng J, Lu H X, He X, et al. Mutation of Spirulina sp. by nuclear irradiation to improve growth rate under 15% carbon dioxide in flue gas[J]. Bioresource Technology, 2017, 238: 650-656. |
| 22 | Zhu Y X, Cheng J, Zhang Z, et al. Mutation of Arthrospira platensis by gamma irradiation to promote phenol tolerance and CO2 fixation for coal-chemical flue gas reduction[J]. Journal of CO2 Utilization, 2020, 38: 252-261. |
| 23 | An J, Gao F L, Ma Q Y, et al. Screening for enhanced astaxanthin accumulation among Spirulina platensis mutants generated by atmospheric and room temperature plasmas[J]. Algal Research, 2017, 25: 464-472. |
| 24 | Fujisawa T, Narikawa R, Okamoto S, et al. Genomic structure of an economically important cyanobacterium, Arthrospira (Spirulina) platensis NIES-39[J]. DNA Research, 2010, 17(2): 85-103. |
| 25 | Cheng J, Lu H X, Li K, et al. Enhancing growth-relevant metabolic pathways of Arthrospira platensis (CYA-1) with gamma irradiation from 60Co[J]. RSC Advances, 2018, 8(30): 16824-16833. |
| 26 | Kurdrid P, Senachak J, Sirijuntarut M, et al. Comparative analysis of the Spirulina platensis subcellular proteome in response to low- and high-temperature stresses: uncovering cross-talk of signaling components[J]. Proteome Sci., 2011, 9: 39. |
| 27 | Depraetere O, Deschoenmaeker F, Badri H, et al. Trade-off between growth and carbohydrate accumulation in nutrient-limited Arthrospira sp. PCC 8005 studied by integrating transcriptomic and proteomic approaches[J]. PLoS One, 2015, 10(7): e0132461. |
| 28 | Kumaresan V, Nizam F, Ravichandran G, et al. Transcriptome changes of blue-green algae, Arthrospira sp. in response to sulfate stress[J]. Algal Research, 2017, 23: 96-103. |
| 29 | Ismaiel M M S, Piercey-Normore M D, Rampitsch C. Proteomic analyses of the cyanobacterium Arthrospira (Spirulina) platensis under iron and salinity stress[J]. Environmental and Experimental Botany, 2018, 147: 63-74. |
| 30 | Tan Y, Fang M Y, Jin L H, et al. Culture characteristics of the atmospheric and room temperature plasma-mutated Spirulina platensis mutants in CO2 aeration culture system for biomass production[J]. Journal of Bioscience and Bioengineering, 2015, 120(4): 438-443. |
| 31 | Vonshak A. Spirulina platensis (Arthrospira): Physiology, Cell Biology and Biotechnology[M]. London: CRC Press, 1997. |
| [1] | 鲁统鹏, 潘晓林, 吴鸿飞, 李煜, 于海燕. 有机絮凝剂对铁矿相沉降性能影响及其吸附机理[J]. 化工学报, 2022, 73(9): 4122-4132. |
| [2] | 李彩风, 王晓, 李岗建, 林军章, 汪卫东, 束青林, 曹嫣镔, 肖盟. 嗜烃乳化菌SL-1与内源菌协同驱油的菌群作用关系研究[J]. 化工学报, 2022, 73(9): 4095-4102. |
| [3] | 张劢, 田瑶, 郭之旗, 王叶, 窦广进, 宋浩. 光催化-生物杂合系统设计优化用于燃料和化学品绿色合成[J]. 化工学报, 2022, 73(7): 2774-2789. |
| [4] | 王靖楠, 庞建, 秦磊, 郭超, 吕波, 李春, 王超. 丁烯基多杀菌素高产菌株的选育和改造策略[J]. 化工学报, 2022, 73(2): 566-576. |
| [5] | 黄明, 朱亮, 丁紫霞, 毛一婷, 马中青. 生物质三组分与低密度聚乙烯共催化热解制取轻质芳烃的协同作用机理[J]. 化工学报, 2022, 73(2): 699-711. |
| [6] | 刘海波, 王楠, 刘洪周, 陈铁柱, 李建昌. 电压扰动对EAD代谢通量中微生物与关键酶活性的影响[J]. 化工学报, 2022, 73(10): 4603-4612. |
| [7] | 侯晓松, 刘晨星, 任爱玲, 郭斌, 郭渊明. 超声雾化/表面活性剂强化吸收耦合生物洗涤净化甲苯废气[J]. 化工学报, 2022, 73(10): 4692-4706. |
| [8] | 宋伟, 王金辉, 胡贵鹏, 陈修来, 刘立明, 吴静. 多酶级联催化合成(R)-β-酪氨酸[J]. 化工学报, 2022, 73(1): 352-361. |
| [9] | 陈婷婷, 韩恺忻, 陈翠雪, 凌雪萍, 沈亮, 卢英华. 铁还原菌Shewanella xiamenensis BC01的有机溶剂应激研究[J]. 化工学报, 2021, 72(7): 3747-3756. |
| [10] | 毛金竹, 肖淑玲, 杨智淳, 王孝宇, 张诗, 陈俊宏, 谢佶晟, 陈福德, 黄子诺, 冯天宇, 张瑷珲, 方柏山. 合成生物学在农残检测领域的应用[J]. 化工学报, 2021, 72(5): 2413-2425. |
| [11] | 赵贞尧, 张保财, 李锋, 宋浩. 产电细胞的合成生物学设计构建[J]. 化工学报, 2021, 72(1): 468-482. |
| [12] | 王凯峰, 王金鹏, 韦萍, 纪晓俊. 代谢工程改造解脂耶氏酵母生产脂肪酸及其衍生物[J]. 化工学报, 2021, 72(1): 351-365. |
| [13] | 秦磊, 俞杰, 宁小钰, 孙文涛, 李春. 合成生物系统构建与绿色生物“智”造[J]. 化工学报, 2020, 71(9): 3979-3994. |
| [14] | 姜岩, 张哲. 不同亲水特性VOCs在生物滴滤工艺中的作用规律[J]. 化工学报, 2020, 71(7): 2973-2982. |
| [15] | 宋易航, 王楚浩, 方柏山. 胶原酶研究进展与应用[J]. 化工学报, 2019, 70(9): 3213-3227. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备 11010102001995号
京公网安备 11010102001995号