CIESC Journal ›› 2025, Vol. 76 ›› Issue (8): 3789-3804.DOI: 10.11949/0438-1157.20250043
• Reviews and monographs • Previous Articles Next Articles
Zhihong CHEN( ), Jiawei WU, Xiaoling LOU, Junxian YUN(
), Jiawei WU, Xiaoling LOU, Junxian YUN( )
)
Received:2025-01-10
Revised:2025-03-10
Online:2025-09-17
Published:2025-08-25
Contact:
Junxian YUN
通讯作者:
贠军贤
作者简介:陈治宏(1997—),男,博士研究生,1471913055@qq.com
基金资助:CLC Number:
Zhihong CHEN, Jiawei WU, Xiaoling LOU, Junxian YUN. Recent advances in machine learning for biomanufacturing of chemicals[J]. CIESC Journal, 2025, 76(8): 3789-3804.
陈治宏, 吴佳伟, 楼小玲, 贠军贤. 化学品生物制造过程机器学习的研究进展[J]. 化工学报, 2025, 76(8): 3789-3804.
Add to citation manager EndNote|Ris|BibTeX
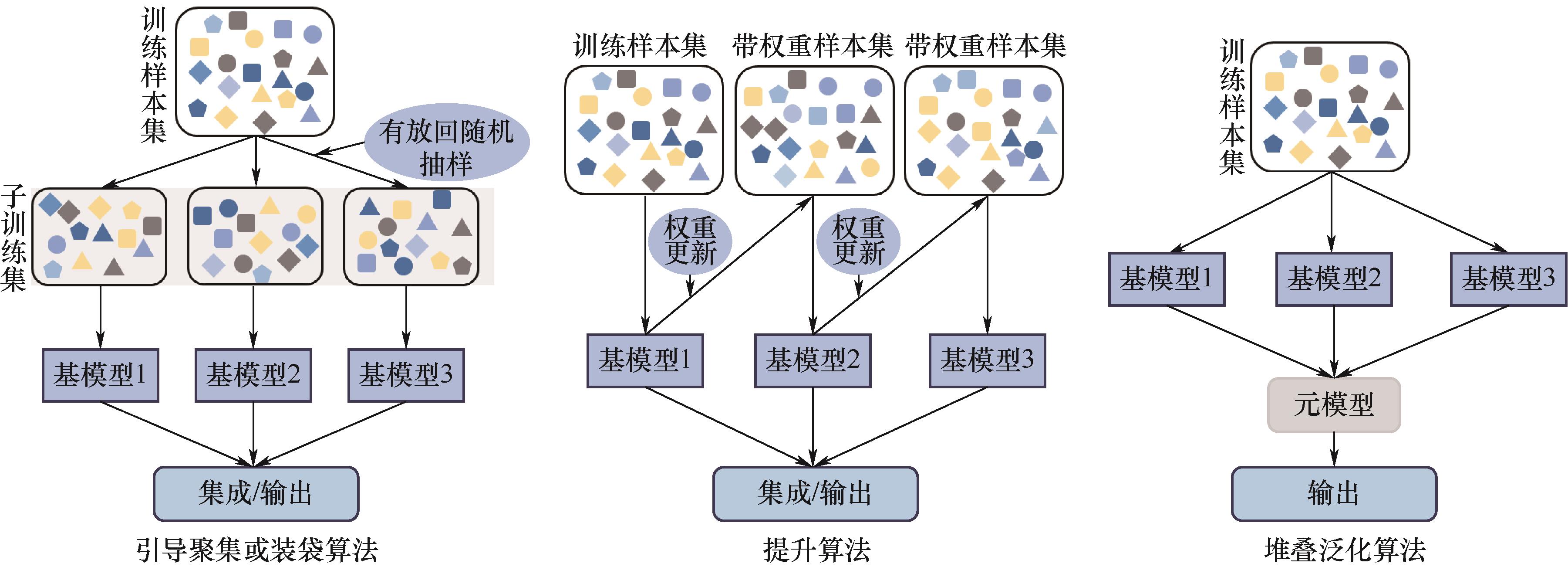
Fig.5 Schematic diagrams of ensemble learning(The base model is the weak learner in ensemble learning, which is the basic unit obtained by training different samples. Weight updating means that the model updates the weight of samples according to the performance of the previous base model, so that the subsequent base model can pay more attention to the difficult-to-classify samples. The meta-model is the final decision model for synthesizing the output of the base model. Integration/output is the output combination of the results of multiple base models according to voting or weighted average to form the final prediction. Symbols with different colors and shapes represent different data input points)
| 应用 | 算法 | 结果 | 文献 |
|---|---|---|---|
| 大肠杆菌发酵生产柠檬烯和红没药烯 | PCA | 产量提高40% | [ |
| 淡水蓝藻培养生产藻胆蛋白 | GSMM、CNN | 产量提高61.76% | [ |
| 黄曲霉发酵生产纤维素酶 | GPR、RBFNN等 | 产量提高3倍 | [ |
| 醋酸菌发酵生产乙酸 | FNN | 产量达1.5~2.0 g/100 ml | [ |
| 嗜热地芽孢杆菌发酵生产脂肪酶 | BPNN | 酶活提高4.7倍 | [ |
| 马克斯克鲁维酵母发酵生产生物乙醇 | ANN | 产量提高至25.4 g/L | [ |
| 微藻培养生产红没药烯 | CNN | 产率提高22% | [ |
Table 1 Applications of machine learning in the optimization of biosynthesis processes
| 应用 | 算法 | 结果 | 文献 |
|---|---|---|---|
| 大肠杆菌发酵生产柠檬烯和红没药烯 | PCA | 产量提高40% | [ |
| 淡水蓝藻培养生产藻胆蛋白 | GSMM、CNN | 产量提高61.76% | [ |
| 黄曲霉发酵生产纤维素酶 | GPR、RBFNN等 | 产量提高3倍 | [ |
| 醋酸菌发酵生产乙酸 | FNN | 产量达1.5~2.0 g/100 ml | [ |
| 嗜热地芽孢杆菌发酵生产脂肪酶 | BPNN | 酶活提高4.7倍 | [ |
| 马克斯克鲁维酵母发酵生产生物乙醇 | ANN | 产量提高至25.4 g/L | [ |
| 微藻培养生产红没药烯 | CNN | 产率提高22% | [ |
| 应用 | 算法 | 结果 | 文献 |
|---|---|---|---|
| 生物反应器泡沫传感器 | CNN | 识别准确率达98% | [ |
| 毕赤酵母发酵生产乙肝表面抗原 | FNN | R2: 0.969;RMSE: 2.717 | [ |
| 食物垃圾厌氧消化 | CNN-BdLSTM | R2: 0.978;RMSE: 0.031 | [ |
| 发酵生产链激酶和青霉素 | DNN | RMSE: 0.0274~0.0565 | [ |
| 乳酸菌发酵生产奶油奶酪 | LSTM | R2: 0.99 | [ |
| 微藻培养生产叶黄素 | ANN | 平均误差为2.6%~11.7% | [ |
Table 2 Applications of machine learning in the monitoring and control of biosynthesis processes
| 应用 | 算法 | 结果 | 文献 |
|---|---|---|---|
| 生物反应器泡沫传感器 | CNN | 识别准确率达98% | [ |
| 毕赤酵母发酵生产乙肝表面抗原 | FNN | R2: 0.969;RMSE: 2.717 | [ |
| 食物垃圾厌氧消化 | CNN-BdLSTM | R2: 0.978;RMSE: 0.031 | [ |
| 发酵生产链激酶和青霉素 | DNN | RMSE: 0.0274~0.0565 | [ |
| 乳酸菌发酵生产奶油奶酪 | LSTM | R2: 0.99 | [ |
| 微藻培养生产叶黄素 | ANN | 平均误差为2.6%~11.7% | [ |
| 应用 | 算法 | 结果 | 文献 |
|---|---|---|---|
| 微滤分离酵母悬液 | ANN | 平均误差小于10% | [ |
| 超滤分离牛血清白蛋白 | ANN | 平均误差小于2.7% | [ |
| 超滤分离苹果皮多酚 | SVM、BPNN | R2: 0.9997;RMSE: 0.03894 | [ |
| 离子对色谱分离寡核苷酸 | SVM | RMSE < 0.2 | [ |
| 反相液相色谱分离肽 | SVM | 平均误差为2.0%~4.2% | [ |
| 混合模式树脂分离蛋白 | RF、GBDT | R2: 0.79~0.82 | [ |
| 蛋白质A层析分离单克隆抗体 | PCA-IF、LSTM等 | R2: 0.96;RMSE: 0.014 | [ |
Table 3 Applications of machine learning in the development of bioseparation processes
| 应用 | 算法 | 结果 | 文献 |
|---|---|---|---|
| 微滤分离酵母悬液 | ANN | 平均误差小于10% | [ |
| 超滤分离牛血清白蛋白 | ANN | 平均误差小于2.7% | [ |
| 超滤分离苹果皮多酚 | SVM、BPNN | R2: 0.9997;RMSE: 0.03894 | [ |
| 离子对色谱分离寡核苷酸 | SVM | RMSE < 0.2 | [ |
| 反相液相色谱分离肽 | SVM | 平均误差为2.0%~4.2% | [ |
| 混合模式树脂分离蛋白 | RF、GBDT | R2: 0.79~0.82 | [ |
| 蛋白质A层析分离单克隆抗体 | PCA-IF、LSTM等 | R2: 0.96;RMSE: 0.014 | [ |
| 应用 | 算法 | 结果 | 文献 |
|---|---|---|---|
| 厌氧消化生产甲烷 | KNN | R2: 0.75 | [ |
| SVM | R2: 0.92;RMSE: 0.167 | [ | |
| 发酵生产生物乙醇 | FNN | R2: 0.91 | [ |
| 酯交换生产生物柴油 | KNN、RF | RMSE: 5.178 | [ |
| Huber-Adaboost、DT-Adaboost | R2: 0.996;RMSE: 1.82 | [ |
Table 4 Applications of machine learning in the production of biofuel chemicals
| 应用 | 算法 | 结果 | 文献 |
|---|---|---|---|
| 厌氧消化生产甲烷 | KNN | R2: 0.75 | [ |
| SVM | R2: 0.92;RMSE: 0.167 | [ | |
| 发酵生产生物乙醇 | FNN | R2: 0.91 | [ |
| 酯交换生产生物柴油 | KNN、RF | RMSE: 5.178 | [ |
| Huber-Adaboost、DT-Adaboost | R2: 0.996;RMSE: 1.82 | [ |
| [1] | Yan X, Liu X, Zhao C H, et al. Applications of synthetic biology in medical and pharmaceutical fields[J]. Signal Transduction and Targeted Therapy, 2023, 8(1): 199. |
| [2] | Tramontina R, Ciancaglini I, Roman E K B, et al. Sustainable biosynthetic pathways to value-added bioproducts from hydroxycinnamic acids[J]. Applied Microbiology and Biotechnology, 2023, 107(13): 4165-4185. |
| [3] | Nielsen J, Keasling J D. Engineering cellular metabolism[J]. Cell, 2016, 164(6): 1185-1197. |
| [4] | Campana F, Brufani G, Mauriello F, et al. Green polyurethanes from bio-based building blocks: recent advances and applications[J]. Green Synthesis and Catalysis, 2024.doi:10.1016/j.gresc.2024.08.001 . |
| [5] | Kang M K, Yoon S H, Kwon M, et al. Microbial cell factories for bio-based isoprenoid production to replace fossil resources[J]. Current Opinion in Systems Biology, 2024, 37: 100502. |
| [6] | Jiang L H, Dong C, Huang L, et al. Metabolic engineering tools for Saccharomyces cerevisiae [J]. Chinese Journal of Biotechnology, 2021, 37(5): 1578-1602. |
| [7] | Mowbray M, Savage T, Wu C F, et al. Machine learning for biochemical engineering: a review[J]. Biochemical Engineering Journal, 2021, 172: 108054. |
| [8] | Kumar Sharma A, Kumar Ghodke P, Goyal N, et al. Machine learning technology in biohydrogen production from agriculture waste: recent advances and future perspectives[J]. Bioresource Technology, 2022, 364: 128076. |
| [9] | Ai Y H, He F, Lancaster E, et al. Application of machine learning for multi-community COVID-19 outbreak predictions with wastewater surveillance[J]. PLoS One, 2022, 17(11): e0277154. |
| [10] | Jabarivelisdeh B, Carius L, Findeisen R, et al. Adaptive predictive control of bioprocesses with constraint-based modeling and estimation[J]. Computers & Chemical Engineering, 2020, 135: 106744. |
| [11] | Gani R, Bałdyga J, Biscans B, et al. A multi-layered view of chemical and biochemical engineering[J]. Chemical Engineering Research and Design, 2020, 155: A133-A145. |
| [12] | Oliveira A L. Biotechnology, big data and artificial intelligence[J]. Biotechnology Journal, 2019, 14(8): 1800613. |
| [13] | Yang C T, Kristiani E, Leong Y K, et al. Big data and machine learning driven bioprocessing—recent trends and critical analysis[J]. Bioresource Technology, 2023, 372: 128625. |
| [14] | Duong-Trung N, Born S, Kim J W, et al. When bioprocess engineering meets machine learning: a survey from the perspective of automated bioprocess development[J]. Biochemical Engineering Journal, 2023, 190: 108764. |
| [15] | Handelman G S, Kok H K, Chandra R V, et al. eDoctor: machine learning and the future of medicine[J]. Journal of Internal Medicine, 2018, 284(6): 603-619. |
| [16] | Gupta V, Mishra V K, Singhal P, et al. An overview of supervised machine learning algorithm[C]//11th International Conference on System Modeling & Advancement in Research Trends (SMART). IEEE, 2022: 87-92. |
| [17] | Hierons R. Machine learning[J]. Software Testing, Verification and Reliability, 1999, 9(3): 191-193. |
| [18] | Jablonka K M, Ongari D, Moosavi S M, et al. Big-data science in porous materials: materials genomics and machine learning[J]. Chemical Reviews, 2020, 120(16): 8066-8129. |
| [19] | Wang M W, Cui Y, Wang X, et al. Machine learning for networking: workflow, advances and opportunities[J]. IEEE Network, 2018, 32(2): 92-99. |
| [20] | Zhong S F, Zhang K, Bagheri M, et al. Machine learning: new ideas and tools in environmental science and engineering[J]. Environmental Science & Technology, 2021, 55(19): 12741-12754. |
| [21] | Wang X L, Zhou G Z, Liang L P, et al. Deep learning-based image analysis for in situ microscopic imaging of cell culture process[J]. Engineering Applications of Artificial Intelligence, 2024, 129: 107621. |
| [22] | Li B, Lin Y Z, Yu W, et al. Application of mechanistic modelling and machine learning for cream cheese fermentation pH prediction[J]. Journal of Chemical Technology & Biotechnology, 2021, 96(1): 125-133. |
| [23] | Wang L G, Long F, Liao W, et al. Prediction of anaerobic digestion performance and identification of critical operational parameters using machine learning algorithms[J]. Bioresource Technology, 2020, 298: 122495. |
| [24] | Zhang L H, Chao B, Zhang X. Modeling and optimization of microbial lipid fermentation from cellulosic ethanol wastewater by Rhodotorula glutinis based on the support vector machine[J]. Bioresource Technology, 2020, 301: 122781. |
| [25] | Seo K W, Seo J, Kim K, et al. Prediction of biogas production rate from dry anaerobic digestion of food waste: process-based approach vs. recurrent neural network black-box model[J]. Bioresource Technology, 2021, 341: 125829. |
| [26] | Zhang A H, Zhu K Y, Zhuang X Y, et al. A robust soft sensor to monitor 1,3-propanediol fermentation process by Clostridium butyricum based on artificial neural network[J]. Biotechnology and Bioengineering, 2020, 117(11): 3345-3355. |
| [27] | Xu L, Liang Y, Huang W E, et al. Rapid detection of six Oceanobacillus species in Daqu starter using single-cell Raman spectroscopy combined with machine learning[J]. Microbial Biotechnology, 2024, 17(2): e14416. |
| [28] | Jordan M I, Mitchell T M. Machine learning: trends, perspectives, and prospects[J]. Science, 2015, 349(6245): 255-260. |
| [29] | Uddin S, Khan A, Hossain M E, et al. Comparing different supervised machine learning algorithms for disease prediction[J]. BMC Medical Informatics and Decision Making, 2019, 19(1): 281. |
| [30] | Glielmo A, Husic B E, Rodriguez A, et al. Unsupervised learning methods for molecular simulation data[J]. Chemical Reviews, 2021, 121(16): 9722-9758. |
| [31] | Rives A, Meier J, Sercu T, et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences[J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(15): e2016239118. |
| [32] | Mondal P P, Galodha A, Verma V K, et al. Review on machine learning-based bioprocess optimization, monitoring, and control systems[J]. Bioresource Technology, 2023, 370: 128523. |
| [33] | Volk M J, Lourentzou I, Mishra S, et al. Biosystems design by machine learning[J]. ACS Synthetic Biology, 2020, 9(7): 1514-1533. |
| [34] | Kiran B R, Sobh I, Talpaert V, et al. Deep reinforcement learning for autonomous driving: a survey[J]. IEEE Transactions on Intelligent Transportation Systems, 2022, 23(6): 4909-4926. |
| [35] | Mnih V, Kavukcuoglu K, Silver D, et al. Human-level control through deep reinforcement learning[J]. Nature, 2015, 518(7540): 529-533. |
| [36] | Lutz I D, Wang S Z, Norn C, et al. Top-down design of protein architectures with reinforcement learning[J]. Science, 2023, 380(6642): 266-273. |
| [37] | Gopakumar V, Tiwari S, Rahman I. A deep learning based data driven soft sensor for bioprocesses[J]. Biochemical Engineering Journal, 2018, 136: 28-39. |
| [38] | Bansal M, Goyal A, Choudhary A. A comparative analysis of K-nearest neighbor, genetic, support vector machine, decision tree, and long short term memory algorithms in machine learning[J]. Decision Analytics Journal, 2022, 3: 100071. |
| [39] | Cortes C, Vapnik V. Support-vector networks[J]. Machine Learning, 1995, 20(3): 273-297. |
| [40] | Wu S G, Wang Y X, Jiang W, et al. Rapid prediction of bacterial heterotrophic fluxomics using machine learning and constraint programming[J]. PLoS Computational Biology, 2016, 12(4): e1004838. |
| [41] | Shawe-Taylor J, Sun S L. A review of optimization methodologies in support vector machines[J]. Neurocomputing, 2011, 74(17): 3609-3618. |
| [42] | Ali L, Wajahat I, Amiri Golilarz N, et al. LDA-GA-SVM: improved hepatocellular carcinoma prediction through dimensionality reduction and genetically optimized support vector machine[J]. Neural Computing and Applications, 2021, 33(7): 2783-2792. |
| [43] | Dong C Y, Chen J. Optimization of process parameters for anaerobic fermentation of corn stalk based on least squares support vector machine[J]. Bioresource Technology, 2019, 271: 174-181. |
| [44] | Okwuashi O, Ndehedehe C E. Deep support vector machine for hyperspectral image classification[J]. Pattern Recognition, 2020, 103: 107298. |
| [45] | Xu A K, Chang H M, Xu Y J, et al. Applying artificial neural networks (ANNs) to solve solid waste-related issues: a critical review[J]. Waste Management, 2021, 124: 385-402. |
| [46] | Krogh A. What are artificial neural networks?[J]. Nature Biotechnology, 2008, 26(2): 195-197. |
| [47] | Bhagya Raj G V S, Dash K K. Comprehensive study on applications of artificial neural network in food process modeling[J]. Critical Reviews in Food Science and Nutrition, 2022, 62(10): 2756-2783. |
| [48] | Chauhan D, Yadav A, Neri F. A multi-agent optimization algorithm and its application to training multilayer perceptron models[J]. Evolving Systems, 2024, 15(3): 849-879. |
| [49] | Zhu Z H, Ye Z F, Tang Y. Nondestructive identification for gender of chicken eggs based on GA-BPNN with double hidden layers[J]. Journal of Applied Poultry Research, 2021, 30(4): 100203. |
| [50] | Konakoglu B. Prediction of geodetic point velocity using MLPNN, GRNN, and RBFNN models: a comparative study[J]. Acta Geodaetica et Geophysica, 2021, 56(2): 271-291. |
| [51] | Dehdarinejad E, Bayareh M. Performance analysis of a novel cyclone separator using RBFNN and MOPSO algorithms[J]. Powder Technology, 2023, 426: 118663. |
| [52] | Wu G Z, Kechavarzi C, Li X G, et al. Machine learning models for predicting PAHs bioavailability in compost amended soils[J]. Chemical Engineering Journal, 2013, 223: 747-754. |
| [53] | Hu Z Z, Yuan Y, Li X, et al. Yield prediction of “Thermal-dissolution based carbon enrichment” treatment on biomass wastes through coupled model of artificial neural network and AdaBoost[J]. Bioresource Technology, 2022, 343: 126083. |
| [54] | Krishna V V S V, Pappa N, Rani S P J V. Deep learning based soft sensor for bioprocess application[C]//2021 IEEE Second International Conference on Control, Measurement and Instrumentation (CMI). IEEE, 2021: 155-159. |
| [55] | LeCun Y, Bengio Y, Hinton G. Deep learning[J]. Nature, 2015, 521(7553): 436-444. |
| [56] | Goh G D, Sing S L, Yeong W Y. A review on machine learning in 3D printing: applications, potential, and challenges[J]. Artificial Intelligence Review, 2021, 54(1): 63-94. |
| [57] | Liu Y, Xue J H, Li D X, et al. Image recognition based on lightweight convolutional neural network: recent advances[J]. Image and Vision Computing, 2024, 146: 105037. |
| [58] | Ascher S, Wang X N, Watson I, et al. Interpretable machine learning to model biomass and waste gasification[J]. Bioresource Technology, 2022, 364: 128062. |
| [59] | Kumar V, Singh Aydav P S, Minz S. Multi-view ensemble learning using multi-objective particle swarm optimization for high dimensional data classification[J]. Journal of King Saud University-Computer and Information Sciences, 2022, 34(10): 8523-8537. |
| [60] | Breiman L. Bagging predictors[J]. Machine Learning, 1996, 24(2): 123-140. |
| [61] | Jin X, Li S H, Ye H R, et al. Investigation and optimization of biodiesel production based on multiple machine learning technologies[J]. Fuel, 2023, 348: 128546. |
| [62] | Bapat P M, Wangikar P P. Optimization of rifamycin B fermentation in shake flasks via a machine-learning-based approach[J]. Biotechnology and Bioengineering, 2004, 86(2): 201-208. |
| [63] | Poth M, Magill G, Filgertshofer A, et al. Extensive evaluation of machine learning models and data preprocessings for Raman modeling in bioprocessing[J]. Journal of Raman Spectroscopy, 2022, 53(9): 1580-1591. |
| [64] | Zhuang L P, Tang B, Bin L Y, et al. Performance prediction of an internal-circulation membrane bioreactor based on models comparison and data features analysis[J]. Biochemical Engineering Journal, 2021, 166: 107850. |
| [65] | Bühlmann P, Yu B. Analyzing bagging[J]. The Annals of Statistics, 2002, 30(4): 927-961. |
| [66] | Freund Y, Schapire R E. Experiments with a new boosting algorithm[C]//Proceedings of the Thirteenth International Conference on International Conference on Machine Learning. Morgan Kaufmann, 1996: 148-156. |
| [67] | Mohammed A, Kora R. A comprehensive review on ensemble deep learning: opportunities and challenges[J]. Journal of King Saud University-Computer and Information Sciences, 2023, 35(2): 757-774. |
| [68] | Dong Z X, Bai X P, Xu D C, et al. Machine learning prediction of pyrolytic products of lignocellulosic biomass based on physicochemical characteristics and pyrolysis conditions[J]. Bioresource Technology, 2023, 367: 128182. |
| [69] | Yaqub M, Lee W. Modeling nutrient removal by membrane bioreactor at a sewage treatment plant using machine learning models[J]. Journal of Water Process Engineering, 2022, 46: 102521. |
| [70] | Smyth P, Wolpert D. Stacked density estimation[C]//Proceedings of the 10th International Conference on Neural Information Processing Systems. Cambridge: MIT Press, 1997: 668-674. |
| [71] | Xiong Y L, Ye M Q, Wu C R. Cancer classification with a cost-sensitive naive Bayes stacking ensemble[J]. Computational and Mathematical Methods in Medicine, 2021, 2021(1): 5556992. |
| [72] | Alonso-Gutierrez J, Kim E M, Batth T S, et al. Principal component analysis of proteomics (PCAP) as a tool to direct metabolic engineering[J]. Metabolic Engineering, 2015, 28: 123-133. |
| [73] | Saini D K, Rai A, Devi A, et al. A multi-objective hybrid machine learning approach-based optimization for enhanced biomass and bioactive phycobiliproteins production in Nostoc sp. CCC-403[J]. Bioresource Technology, 2021, 329: 124908. |
| [74] | Singh V, Haque S, Niwas R, et al. Strategies for fermentation medium optimization: an in-depth review[J]. Frontiers in Microbiology, 2017, 7: 2087. |
| [75] | Witek-Krowiak A, Chojnacka K, Podstawczyk D, et al. Application of response surface methodology and artificial neural network methods in modelling and optimization of biosorption process[J]. Bioresource Technology, 2014, 160: 150-160. |
| [76] | Singhal A, Kumari N, Ghosh P, et al. Optimizing cellulase production from Aspergillus flavus using response surface methodology and machine learning models[J]. Environmental Technology & Innovation, 2022, 27: 102805. |
| [77] | Upadhyay A, Kovalev A A, Zhuravleva E A, et al. Enhanced production of acetic acid through bioprocess optimization employing response surface methodology and artificial neural network[J]. Bioresource Technology, 2023, 376: 128930. |
| [78] | Ebrahimpour A, Abd Rahman R N Z, Ean Ch'ng D H, et al. A modeling study by response surface methodology and artificial neural network on culture parameters optimization for thermostable lipase production from a newly isolated thermophilic Geobacillus sp. strain ARM[J]. BMC Biotechnology, 2008, 8: 96. |
| [79] | Elumalai R S, Ramanujam P, Tawfik M A, et al. Optimization and kinetics modelling for enhancing the bioethanol production from banana peduncle using Trichoderma reesei and Kluveromyces marxianus by co-pretreatment methods[J]. Sustainable Energy Technologies and Assessments, 2023, 56: 103129. |
| [80] | Rajasekhar N, Radhakrishnan T K, Mohamed S N. Reinforcement learning based temperature control of a fermentation bioreactor for ethanol production[J]. Biotechnology and Bioengineering, 2024, 121(10): 3114-3127. |
| [81] | Chai W Y, Tan M K, Teo K T K, et al. Optimization of fed-batch baker's yeast fermentation using deep reinforcement learning[J]. Process Integration and Optimization for Sustainability, 2024, 8(2): 395-411. |
| [82] | del Rio-Chanona E A, Wagner J L, Ali H, et al. Deep learning-based surrogate modeling and optimization for microalgal biofuel production and photobioreactor design[J]. AIChE Journal, 2019, 65(3): 915-923. |
| [83] | Takors R. Scale-up of microbial processes: impacts, tools and open questions[J]. Journal of Biotechnology, 2012, 160(1/2): 3-9. |
| [84] | Bayer B, von Stosch M, Striedner G, et al. Comparison of modeling methods for DoE-based holistic upstream process characterization[J]. Biotechnology Journal, 2020, 15(5): 1900551. |
| [85] | Austerjost J, Söldner R, Edlund C, et al. A machine vision approach for bioreactor foam sensing[J]. SLAS Technology, 2021, 26(4): 408-414. |
| [86] | Zhu X L, Rehman K U, Wang B, et al. Modern soft-sensing modeling methods for fermentation processes[J]. Sensors, 2020, 20(6): 1771. |
| [87] | Hosseini S N, Javidanbardan A, Khatami M. Accurate and cost-effective prediction of HBsAg titer in industrial scale fermentation process of recombinant Pichia pastoris by using neural network based soft sensor[J]. Biotechnology and Applied Biochemistry, 2019, 66(4): 681-689. |
| [88] | Kazemi P, Steyer J P, Bengoa C, et al. Robust data-driven soft sensors for online monitoring of volatile fatty acids in anaerobic digestion processes[J]. Processes, 2020, 8(1): 67. |
| [89] | Jia R, Song Y C, Piao D M, et al. Exploration of deep learning models for real-time monitoring of state and performance of anaerobic digestion with online sensors[J]. Bioresource Technology, 2022, 363: 127908. |
| [90] | Liu Y Q, Zhu Z Y, Zhu X L. Soft sensor modeling for key parameters of marine alkaline protease MP fermentation process[C]//2018 Chinese Control and Decision Conference (CCDC). IEEE, 2018: 6149-6154. |
| [91] | Zhang D D, Del Rio-Chanona E A, Petsagkourakis P, et al. Hybrid physics-based and data-driven modeling for bioprocess online simulation and optimization[J]. Biotechnology and Bioengineering, 2019, 116(11): 2919-2930. |
| [92] | Singh P C, Singh R K. Choosing an appropriate bioseparation technique[J]. Trends in Food Science & Technology, 1996, 7(2): 49-58. |
| [93] | Keller K, Friedmann T, Boxman A. The bioseparation needs for tomorrow[J]. Trends in Biotechnology, 2001, 19(11): 438-441. |
| [94] | Orr V, Zhong L Y, Moo-Young M, et al. Recent advances in bioprocessing application of membrane chromatography[J]. Biotechnology Advances, 2013, 31(4): 450-465. |
| [95] | Hemida M, Haidar Ahmad I A, Barrientos R C, et al. Computer-assisted multifactorial method development for the streamlined separation and analysis of multicomponent mixtures in (Bio)pharmaceutical settings[J]. Analytica Chimica Acta, 2024, 1293: 342178. |
| [96] | Poole C, Cabooter D. Editorial: special issue machine learning and other tools for data handling in chromatography[J]. Journal of Chromatography A, 2022, 1684: 463579. |
| [97] | Rizki Z, Ottens M. Model-based optimization approaches for pressure-driven membrane systems[J]. Separation and Purification Technology, 2023, 315: 123682. |
| [98] | Ní Mhurchú J, Foley G. Dead-end filtration of yeast suspensions: correlating specific resistance and flux data using artificial neural networks[J]. Journal of Membrane Science, 2006, 281(1/2): 325-333. |
| [99] | Bowen W R, Jones M G, Yousef H N S. Dynamic ultrafiltration of proteins–a neural network approach[J]. Journal of Membrane Science, 1998, 146(2): 225-235. |
| [100] | Park S, Baek S S, Pyo J, et al. Deep neural networks for modeling fouling growth and flux decline during NF/RO membrane filtration[J]. Journal of Membrane Science, 2019, 587: 117164. |
| [101] | Wang L, Li Z H, Fan J H, et al. Prediction of membrane purification by membrane fouling based on mathematic and machine learning models combined with image processing technology[J]. Journal of Environmental Chemical Engineering, 2023, 11(5): 111154. |
| [102] | Yun J X, Cheng X H, Ye J L, et al. Chromatographic adsorption of serum albumin and antibody proteins in cryogels with benzyl-quaternary amine ligands[J]. Journal of Chromatography A, 2015, 1381: 173-183. |
| [103] | Cramer S M, Jayaraman G. Preparative chromatography in biotechnology[J]. Current Opinion in Biotechnology, 1993, 4(2): 217-225. |
| [104] | Enmark M, Häggström J, Samuelsson J, et al. Building machine-learning-based models for retention time and resolution predictions in ion pair chromatography of oligonucleotides[J]. Journal of Chromatography A, 2022, 1671: 462999. |
| [105] | Samuelsson J, Eiriksson F F, Dennis A, et al. Determining gradient conditions for peptide purification in RPLC with machine-learning-based retention time predictions[J]. Journal of Chromatography A, 2019, 1598: 92-100. |
| [106] | Jäpel R C, Buyel J F. Bayesian optimization using multiple directional objective functions allows the rapid inverse fitting of parameters for chromatography simulations[J]. Journal of Chromatography A, 2022, 1679: 463408. |
| [107] | Kensert A, Bosten E, Collaerts G, et al. Convolutional neural network for automated peak detection in reversed-phase liquid chromatography[J]. Journal of Chromatography A, 2022, 1672: 463005. |
| [108] | 乔红梅. 聚合物型色谱填料的研究进展[J]. 材料科学与工程学报, 2016, 34(2): 333-337. |
| Qiao H M. Progress of polymeric chromatographic packing[J]. Journal of Materials Science and Engineering, 2016, 34(2): 333-337. | |
| [109] | Cai Q Y, Qiao L Z, Yao S J, et al. Machine learning assisted QSAR analysis to predict protein adsorption capacities on mixed-mode resins[J]. Separation and Purification Technology, 2024, 340: 126762. |
| [110] | Mattiasson B, Kumar A, Galaev I Y. Macroporous Polymers: Production Properties and Biotechnological/biomedical Applications[M]. Boca Raton: CRC Press, 2009: 200-205. |
| [111] | Plieva F M, Galaev I Y, Noppe W, et al. Cryogel applications in microbiology[J]. Trends in Microbiology, 2008, 16(11): 543-551. |
| [112] | Lozinsky V I. Cryogels on the basis of natural and synthetic polymers: preparation, properties and applications[J]. Russian Chemical Reviews, 2002, 71(6): 489-511. |
| [113] | Lozinsky V I, Galaev I Y, Plieva F M, et al. Polymeric cryogels as promising materials of biotechnological interest[J]. Trends in Biotechnology, 2003, 21(10): 445-451. |
| [114] | Yun J X, Shen S C, Chen F, et al. One-step isolation of adenosine triphosphate from crude fermentation broth of Saccharomyces cerevisiae by anion-exchange chromatography using supermacroporous cryogel[J]. Journal of Chromatography B, 2007, 860(1): 57-62. |
| [115] | Wang L H, Shen S C, Yun J X, et al. Chromatographic separation of cytidine triphosphate from fermentation broth of yeast using anion-exchange cryogel[J]. Journal of Separation Science, 2008, 31(4): 689-695. |
| [116] | Perçin I, Sağlar E, Yavuz H, et al. Poly(hydroxyethyl methacrylate) based affinity cryogel for plasmid DNA purification[J]. International Journal of Biological Macromolecules, 2011, 48(4): 577-582. |
| [117] | Pan M M, Shen S C, Chen L, et al. Separation of lactoperoxidase from bovine whey milk by cation exchange composite cryogel embedded macroporous cellulose beads[J]. Separation and Purification Technology, 2015, 147: 132-138. |
| [118] | Guan J T, Guan Y X, Yun J X, et al. Chromatographic separation of phenyllactic acid from crude broth using cryogels with dual functional groups[J]. Journal of Chromatography A, 2018, 1554: 92-100. |
| [119] | Wu J W, Wang R B, Tan Y, et al. Hybrid machine learning model based predictions for properties of poly(2-hydroxyethyl methacrylate)-poly(vinyl alcohol) composite cryogels embedded with bacterial cellulose[J]. Journal of Chromatography A, 2024, 1727: 464996. |
| [120] | Tiwari A, Bansode V, Rathore A S. Application of advanced machine learning algorithms for anomaly detection and quantitative prediction in protein A chromatography[J]. Journal of Chromatography A, 2022, 1682: 463486. |
| [121] | Guo M X, Song W P, Buhain J. Bioenergy and biofuels: history, status, and perspective[J]. Renewable and Sustainable Energy Reviews, 2015, 42: 712-725. |
| [122] | Liao M C, Yao Y. Applications of artificial intelligence-based modeling for bioenergy systems: a review[J]. GCB Bioenergy, 2021, 13(5): 774-802. |
| [123] | Wang Z X, Peng X G, Xia A, et al. Comparison of machine learning methods for predicting the methane production from anaerobic digestion of lignocellulosic biomass[J]. Energy, 2023, 263: 125883. |
| [124] | Perera S M H D, Wickramasinghe C, Samarasiri B K T, et al. Modeling of thermochemical conversion of waste biomass—a comprehensive review[J]. Biofuel Research Journal, 2021, 8(4): 1481-1528. |
| [125] | Ge Y D, Tao J Y, Wang Z, et al. Modification of anaerobic digestion model No.1 with machine learning models towards applicable and accurate simulation of biomass anaerobic digestion[J]. Chemical Engineering Journal, 2023, 454: 140369. |
| [126] | Pereira R D, Badino A C, Cruz A J G. Framework based on artificial intelligence to increase industrial bioethanol production[J]. Energy & Fuels, 2020, 34(4): 4670-4677. |
| [127] | Almohana A I, Almojil S F, Kamal M A, et al. Theoretical investigation on optimization of biodiesel production using waste cooking oil: machine learning modeling and experimental validation[J]. Energy Reports, 2022, 8: 11938-11951. |
| [1] |
Jichao GUO, Xiaoxiao XU, Yunlong SUN.
Airflow simulation and optimization based on |
| [2] | Xiaoguang MI, Guogang SUN, Hao CHENG, Xiaohui ZHANG. Performance simulation model and validation of printed circuit natural gas cooler [J]. CIESC Journal, 2025, 76(S1): 426-434. |
| [3] | Xiayu FAN, Jianchen SUN, Keying LI, Xinya YAO, Hui SHANG. Machine learning drives system optimization of liquid organic hydrogen storage technology [J]. CIESC Journal, 2025, 76(8): 3805-3821. |
| [4] | Jinghao ZHANG, Yajun WANG, Yongkang ZHANG. Evaluation of chemical process operation status based on NRBO-SLSTM [J]. CIESC Journal, 2025, 76(8): 4145-4154. |
| [5] | Zheng GAO, Hui WANG, Zhiguo QU. Data-driven high-throughput screening of anion-pillared metal-organic frameworks for hydrogen storage [J]. CIESC Journal, 2025, 76(8): 4259-4272. |
| [6] | Xiaolong WU, Xiaohuang HUANG, Yuan XIAO, Linghai SHAN, Jiahui YE, Guomin CUI. Reserve empty node strategy applied to optimization of heat exchanger networks [J]. CIESC Journal, 2025, 76(7): 3388-3402. |
| [7] | Jingxin ZHANG, Jiaojie HE, Qingwang CAI, Ziyi KANG, Yusi YANG, Tong WANG, Xiantao CAO, Liwei YANG. Prediction of COD concentration in wastewater treatment plant effluent based on secondary decomposition and BiLSTM [J]. CIESC Journal, 2025, 76(6): 2859-2871. |
| [8] | Yulun WU, Zhenlei WANG, Xin WANG. Contrastive learning based on method for identifying operating conditions of ethylene cracking furnace [J]. CIESC Journal, 2025, 76(6): 2733-2742. |
| [9] | Fuyu WANG, Xuanyi ZHOU. Leakage estimation in a chemical tank farm with unsteady adjoint equation and genetic algorithm [J]. CIESC Journal, 2025, 76(6): 3104-3114. |
| [10] | Yifei WANG, Jingjie REN, Mingshu BI, Haotian YE. Multi-objective optimization of cyclohexane oxidation process parameters based on inherent safety and economic performance [J]. CIESC Journal, 2025, 76(6): 2722-2732. |
| [11] | Hanchuan ZHANG, Chao SHANG, Wenxiang LYU, Dexiang HUANG, Yaning ZHANG. Operating conditions pattern recognition and yield prediction for FCCU based on unsupervised time series clustering [J]. CIESC Journal, 2025, 76(6): 2781-2790. |
| [12] | Lin LI, Mingmei WANG, Erwei SONG, Wenwen WANG, Yaochang ZHANG, Erqiang WANG. Thermodynamic analysis and optimization of isoprene/n-pentane separation process [J]. CIESC Journal, 2025, 76(6): 2549-2558. |
| [13] | Jialang HU, Mingyuan JIANG, Lyuming JIN, Yonggang ZHANG, Peng HU, Hongbing JI. Machine learning-assisted high-throughput computational screening of MOFs and advances in gas separation research [J]. CIESC Journal, 2025, 76(5): 1973-1996. |
| [14] | Hanxiao ZHANG, Ruiqi WANG, Yating ZHANG. Prediction of scale factor of heat exchangers based on CNN-LSTM neural network [J]. CIESC Journal, 2025, 76(4): 1671-1679. |
| [15] | Wen CHAN, Wan YU, Gang WANG, Huashan SU, Fenxia HUANG, Tao HU. Thermodynamic and economic analyses and dual-objective optimization of Allam cycle with improved regenerator layout [J]. CIESC Journal, 2025, 76(4): 1680-1692. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||