CIESC Journal ›› 2022, Vol. 73 ›› Issue (2): 521-534.DOI: 10.11949/0438-1157.20211164
• Reviews and monographs • Previous Articles Next Articles
Yi SUN1( ),Teng ZHANG1,Bo LYU1(
),Teng ZHANG1,Bo LYU1( ),Chun LI1,2(
),Chun LI1,2( )
)
Received:2021-08-16
Revised:2021-11-17
Online:2022-02-18
Published:2022-02-05
Contact:
Bo LYU,Chun LI
通讯作者:
吕波,李春
作者简介:孙怡(1997—),女,硕士研究生,基金资助:CLC Number:
Yi SUN, Teng ZHANG, Bo LYU, Chun LI. Improvement for fine regulation of microbial cell factory by intracellular biosensors[J]. CIESC Journal, 2022, 73(2): 521-534.
孙怡, 张腾, 吕波, 李春. 胞内生物传感器提高微生物细胞工厂的精细调控[J]. 化工学报, 2022, 73(2): 521-534.
Add to citation manager EndNote|Ris|BibTeX
| 胞内生物传感器的关键元件 | 响应因子 | 微生物细胞宿主 | 文献 | ||
|---|---|---|---|---|---|
| 转录调节元件 | 启动子 | gadE, rstA | 法尼基焦磷酸 | 大肠杆菌 | [ |
| hmgA | 尿黑酸 | 铜绿假单胞菌 | [ | ||
| araBAD | 脱氧紫色杆菌素 | 大肠杆菌 | [ | ||
| srfA | 细胞密度 | 枯草芽孢杆菌 | [ | ||
| gas | 衣康酸 | 黑曲霉 | [ | ||
| hsp12, hsp26 | 高温,乙酸 | 酿酒酵母 | [ | ||
| 转录因子 | FapR | 丙二酰辅酶A | 酿酒酵母 | [ | |
| Lrp | L-缬氨酸 | 谷氨酸棒杆菌 | [ | ||
| AgaR | L-精氨酸 | 钝齿棒杆菌 | [ | ||
| Saro-0803 | 白藜芦醇 | 大肠杆菌 | [ | ||
| 翻译调节元件 | 核糖体开关 | 茶碱RNA适体 | 茶碱 | 大肠杆菌 | [ |
| 菠菜RNA适体 | 硫胺素5′-焦磷酸, S-腺苷-同型半胱氨酸 | 大肠杆菌 | [ | ||
| 荧光RNA适体 | 胍 | 大肠杆菌 | [ | ||
| 蛋白质 | G蛋白偶联受体 | 丁香酚、香豆素、二氢茉莉酮和苯乙酮 | 酿酒酵母 | [ | |
| 双组分系统 | 苹果酸 | 大肠杆菌 | [ | ||
| F?rster共振能量转移系统 | L-2-羟基戊二酸 | 恶臭假单胞菌 | [ | ||
| 酶偶联 | l-3,4-二羟基苯丙氨酸 | 酿酒酵母 | [ | ||
| 木糖 | 酿酒酵母 | [ | |||
| 乳酸 | 大肠杆菌 | [ | |||
Table 1 Classification of intracellular biosensors and related examples
| 胞内生物传感器的关键元件 | 响应因子 | 微生物细胞宿主 | 文献 | ||
|---|---|---|---|---|---|
| 转录调节元件 | 启动子 | gadE, rstA | 法尼基焦磷酸 | 大肠杆菌 | [ |
| hmgA | 尿黑酸 | 铜绿假单胞菌 | [ | ||
| araBAD | 脱氧紫色杆菌素 | 大肠杆菌 | [ | ||
| srfA | 细胞密度 | 枯草芽孢杆菌 | [ | ||
| gas | 衣康酸 | 黑曲霉 | [ | ||
| hsp12, hsp26 | 高温,乙酸 | 酿酒酵母 | [ | ||
| 转录因子 | FapR | 丙二酰辅酶A | 酿酒酵母 | [ | |
| Lrp | L-缬氨酸 | 谷氨酸棒杆菌 | [ | ||
| AgaR | L-精氨酸 | 钝齿棒杆菌 | [ | ||
| Saro-0803 | 白藜芦醇 | 大肠杆菌 | [ | ||
| 翻译调节元件 | 核糖体开关 | 茶碱RNA适体 | 茶碱 | 大肠杆菌 | [ |
| 菠菜RNA适体 | 硫胺素5′-焦磷酸, S-腺苷-同型半胱氨酸 | 大肠杆菌 | [ | ||
| 荧光RNA适体 | 胍 | 大肠杆菌 | [ | ||
| 蛋白质 | G蛋白偶联受体 | 丁香酚、香豆素、二氢茉莉酮和苯乙酮 | 酿酒酵母 | [ | |
| 双组分系统 | 苹果酸 | 大肠杆菌 | [ | ||
| F?rster共振能量转移系统 | L-2-羟基戊二酸 | 恶臭假单胞菌 | [ | ||
| 酶偶联 | l-3,4-二羟基苯丙氨酸 | 酿酒酵母 | [ | ||
| 木糖 | 酿酒酵母 | [ | |||
| 乳酸 | 大肠杆菌 | [ | |||
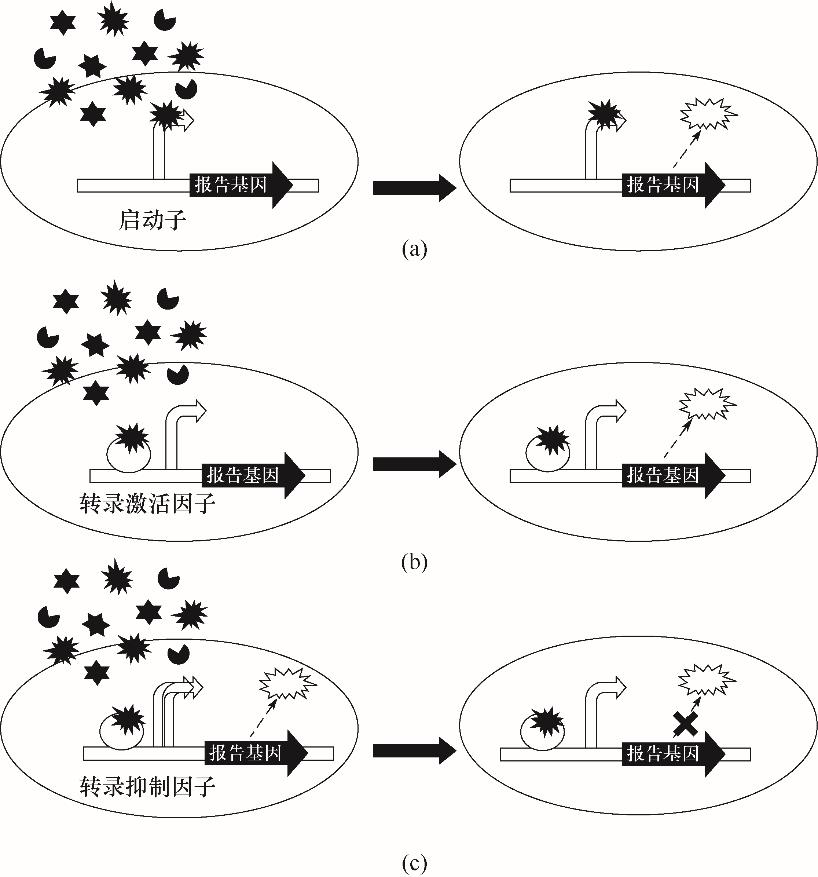
Fig.3 Biosensors based on transcription regulatory elements(a) biosensor based on promoter; (b) biosensor based on transcription activator; (c) biosensor based on transcription inhibitor

Fig.4 Schematic diagram of the “ribosomal switch” biosensors(a) stop transcription by inhibiting anti-terminator; (b) transcribing by isolating ribosome binding site translation; (c) stop transcription by isolating ribosome binding site translation; (d) post-transcription by mRNA cleavage
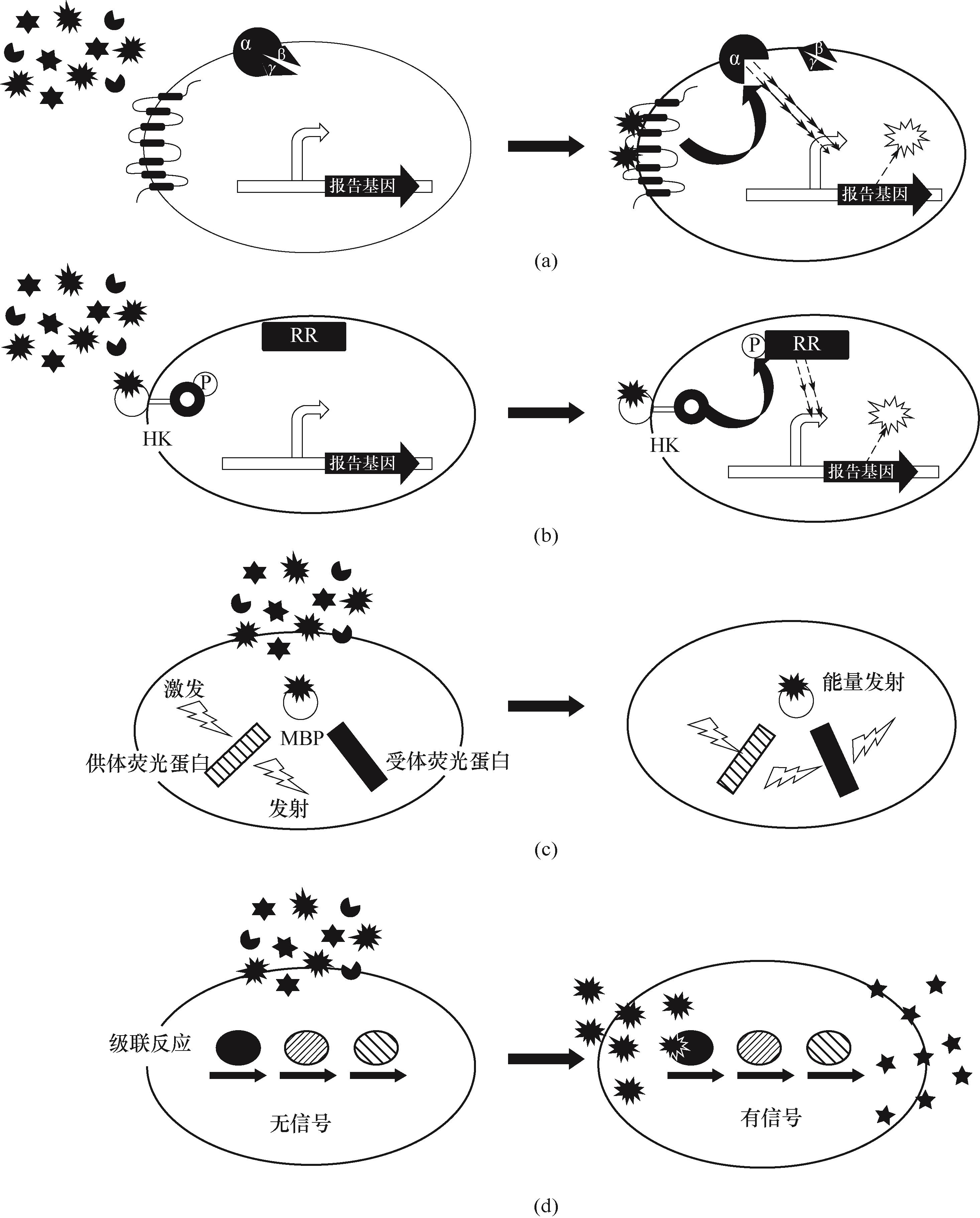
Fig.5 Schematic diagram of protein biosensors work(a) G protein-coupled receptor-based biosensor; (b) two-component system biosensor; (c) F?rster resonance energy transfer biosensor; (d) enzyme-coupled biosensor
| 胞内生物传感器类型 | 关键元件 | 应用目的 | 目标化合物 | 微生物细胞宿主 | 文献 |
|---|---|---|---|---|---|
| 基于转录因子 | ChnR | 高通量筛选优良菌株 | 内酰胺 | 大肠杆菌 | [ |
| 基于转录因子 | FapR | 高通量筛选优良菌株 | 丙二酰辅酶A | 大肠杆菌 | [ |
| 基于转录因子 | XylR | 高通量筛选优良菌株 | 木糖 | 酿酒酵母 | [ |
| 基于转录因子 | SoxR | 高通量筛选优良菌株 | NADPH | 大肠杆菌 | [ |
| 基于转录因子 | C4-lysR | 高通量筛选优良菌株 | 3-羟基丙酸 | 大肠杆菌 | [ |
| 核糖体开关 | glmS | 高通量筛选优良菌株 | 乙酰神经氨酸 | 大肠杆菌 | [ |
| 核糖体开关 | 锤头状核酶 | 高通量筛选优良菌株 | 新霉素 | 酿酒酵母 | [ |
| 核糖体开关 | 色氨酸RNA适体 | 高通量筛选优良菌株 | 色氨酸 | 大肠杆菌 | [ |
| 核糖体开关 | glmS | 高通量筛选优良菌株 | N-乙酰氨基葡萄糖 | 酿酒酵母 | [ |
| 基于转录因子 | FadR | 动态调控代谢平衡 | 香草酸 | 大肠杆菌 | [ |
| 基于转录因子 | IpsA | 动态调控代谢平衡 | D-葡萄糖二酸 | 大肠杆菌 | [ |
| 基于蛋白质 | 双组分系统 | 动态调控代谢平衡 | α-法尼烯 | 酿酒酵母 | [ |
| 基于转录因子 | LuxR,TetR | 动态调控代谢平衡 | 酪醇,红景天苷 | 大肠杆菌 | [ |
| 基于启动子 | AraCmev | 动态调控代谢平衡 | 甲羟戊酸 | 大肠杆菌 | [ |
| 基于转录因子 | FedR,PadR | 动态调控代谢平衡 | 柚皮素 | 大肠杆菌 | [ |
Table 2 Classic cases of intracellular biosensors applied to microbial cell factories
| 胞内生物传感器类型 | 关键元件 | 应用目的 | 目标化合物 | 微生物细胞宿主 | 文献 |
|---|---|---|---|---|---|
| 基于转录因子 | ChnR | 高通量筛选优良菌株 | 内酰胺 | 大肠杆菌 | [ |
| 基于转录因子 | FapR | 高通量筛选优良菌株 | 丙二酰辅酶A | 大肠杆菌 | [ |
| 基于转录因子 | XylR | 高通量筛选优良菌株 | 木糖 | 酿酒酵母 | [ |
| 基于转录因子 | SoxR | 高通量筛选优良菌株 | NADPH | 大肠杆菌 | [ |
| 基于转录因子 | C4-lysR | 高通量筛选优良菌株 | 3-羟基丙酸 | 大肠杆菌 | [ |
| 核糖体开关 | glmS | 高通量筛选优良菌株 | 乙酰神经氨酸 | 大肠杆菌 | [ |
| 核糖体开关 | 锤头状核酶 | 高通量筛选优良菌株 | 新霉素 | 酿酒酵母 | [ |
| 核糖体开关 | 色氨酸RNA适体 | 高通量筛选优良菌株 | 色氨酸 | 大肠杆菌 | [ |
| 核糖体开关 | glmS | 高通量筛选优良菌株 | N-乙酰氨基葡萄糖 | 酿酒酵母 | [ |
| 基于转录因子 | FadR | 动态调控代谢平衡 | 香草酸 | 大肠杆菌 | [ |
| 基于转录因子 | IpsA | 动态调控代谢平衡 | D-葡萄糖二酸 | 大肠杆菌 | [ |
| 基于蛋白质 | 双组分系统 | 动态调控代谢平衡 | α-法尼烯 | 酿酒酵母 | [ |
| 基于转录因子 | LuxR,TetR | 动态调控代谢平衡 | 酪醇,红景天苷 | 大肠杆菌 | [ |
| 基于启动子 | AraCmev | 动态调控代谢平衡 | 甲羟戊酸 | 大肠杆菌 | [ |
| 基于转录因子 | FedR,PadR | 动态调控代谢平衡 | 柚皮素 | 大肠杆菌 | [ |
| 1 | Otero-Muras I, Carbonell P. Automated engineering of synthetic metabolic pathways for efficient biomanufacturing[J]. Metabolic Engineering, 2021, 63: 61-80. |
| 2 | Orsi E, Beekwilder J, Eggink G, et al. The transition of Rhodobacter sphaeroides into a microbial cell factory[J]. Biotechnology and Bioengineering, 2021, 118(2): 531-541. |
| 3 | Yang Y, Lin Y, Li L, et al. Regulating malonyl-CoA metabolism via synthetic antisense RNAs for enhanced biosynthesis of natural products[J]. Metabolic Engineering, 2015, 29: 217-226. |
| 4 | Humphreys C M, Minton N P. Advances in metabolic engineering in the microbial production of fuels and chemicals from C1 gas[J]. Current Opinion in Biotechnology, 2018, 50: 174-181. |
| 5 | Wang J, Zhang R H, Zhang Y, et al. Developing a pyruvate-driven metabolic scenario for growth-coupled microbial production [J]. Metabolic Engineering, 2019, 55: 191-200. |
| 6 | Jiang T, Li C Y, Teng Y X, et al. Recent advances in improving metabolic robustness of microbial cell factories[J]. Current Opinion in Biotechnology, 2020, 66: 69-77. |
| 7 | Fujiwara R, Noda S, Tanaka T, et al. Metabolic engineering of Escherichia coli for shikimate pathway derivative production from glucose-xylose co-substrate[J]. Nature Communications, 2020, 11: 279. |
| 8 | Luo X Z, Reiter M A, D'Espaux L, et al. Complete biosynthesis of cannabinoids and their unnatural analogues in yeast[J]. Nature, 2019, 567(7746): 123-126. |
| 9 | Tian R Z, Liu Y F, Chen J R, et al. Synthetic N-terminal coding sequences for fine-tuning gene expression and metabolic engineering in Bacillus subtilis[J]. Metabolic Engineering, 2019, 55: 131-141. |
| 10 | Yu Y, Chang P, Yu H, et al. Productive amyrin synthases for efficient α-amyrin synthesis in engineered Saccharomyces cerevisiae[J]. ACS Synthetic Biology, 2018, 7(10): 2391-2402. |
| 11 | Yu Y, Rasool A, Liu H, et al. Engineering Saccharomyces cerevisiae for high yield production of α-amyrin via synergistic remodeling of α-amyrin synthase and expanding the storage pool[J]. Metabolic Engineering, 2020, 62: 72-83. |
| 12 | Liu D Y, Jin J, Gao M, et al. Dataset for the estimation of costs for direct contact condenser[J]. Data in Brief, 2018, 20: 535-543. |
| 13 | Jang S, Jang S, Xiu Y, et al. Development of artificial riboswitches for monitoring of naringenin in vivo[J]. ACS Synthetic Biology, 2017, 6(11): 2077-2085. |
| 14 | Purdy H M, Reed J L. Evaluating the capabilities of microbial chemical production using genome-scale metabolic models[J]. Current Opinion in Systems Biology, 2017, 2: 91-97. |
| 15 | Nielsen A A K, Der B S, Shin J, et al. Genetic circuit design automation[J]. Science, 2016, 352(6281): aac7341. |
| 16 | Chae T U, Choi S Y, Kim J W, et al. Recent advances in systems metabolic engineering tools and strategies[J]. Current Opinion in Biotechnology, 2017, 47: 67-82. |
| 17 | Hanko E K R, Minton N P, Malys N. A transcription factor-based biosensor for detection of itaconic acid[J]. ACS Synthetic Biology, 2018, 7(5): 1436-1446. |
| 18 | Ghazi Z, Jahanshahi S, Li Y. RiboFACSeq: a new method for investigating metabolic and transport pathways in bacterial cells by combining a riboswitch-based sensor, fluorescence-activated cell sorting and next-generation sequencing[J]. PLoS One, 2017, 12(12): e0188399. |
| 19 | Kalkreuter E, Keeler A M, Malico A A, et al. Development of a genetically encoded biosensor for detection of polyketide synthase extender units in Escherichia coli[J]. ACS Synthetic Biology, 2019, 8(6): 1391-1400. |
| 20 | Johnson A O, Gonzalez-Villanueva M, Wong L, et al. Design and application of genetically-encoded malonyl-CoA biosensors for metabolic engineering of microbial cell factories[J]. Metabolic Engineering, 2017, 44: 253-264. |
| 21 | Doong S J, Gupta A, Prather K L J. Layered dynamic regulation for improving metabolic pathway productivity in Escherichia coli[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(12): 2964-2969. |
| 22 | de Paepe B, Maertens J, Vanholme B, et al. Modularization and response curve engineering of a naringenin-responsive transcriptional biosensor[J]. ACS Synthetic Biology, 2018, 7(5): 1303-1314. |
| 23 | Liu D, Zhang F. Metabolic feedback circuits provide rapid control of metabolite dynamics[J]. ACS Synthetic Biology, 2018, 7(2): 347-356. |
| 24 | Cevenini L, Calabretta M M, Lopreside A, et al. Exploiting NanoLuc luciferase for smartphone-based bioluminescence cell biosensor for (anti)-inflammatory activity and toxicity[J]. Analytical and Bioanalytical Chemistry, 2016, 408(30): 8859-8868. |
| 25 | Gui Q Y, Lawson T, Shan S Y, et al. The application of whole cell-based biosensors for use in environmental analysis and in medical diagnostics[J]. Sensors, 2017, 17(7): 1623. |
| 26 | Bronder T S, Poghossian A, Scheja S, et al. DNA immobilization and hybridization detection by the intrinsic molecular charge using capacitive field-effect sensors modified with a charged weak polyelectrolyte layer[J]. ACS Applied Materials & Interfaces, 2015, 7(36): 20068-20075. |
| 27 | Ma Q, Zhang Q W, Xu Q Y, et al. Systems metabolic engineering strategies for the production of amino acids[J]. Synthetic and Systems Biotechnology, 2017, 2(2): 87-96. |
| 28 | Yeom S J, Kim M, Kwon K K, et al. A synthetic microbial biosensor for high-throughput screening of lactam biocatalysts[J]. Nature Communications, 2018, 9: 5053. |
| 29 | D'Ambrosio V, Jensen M K. Lighting up yeast cell factories by transcription factor-based biosensors[J]. FEMS Yeast Research, 2017, 17(7): fox076. |
| 30 | Dahl R H, Zhang F Z, Alonso-Gutierrez J, et al. Engineering dynamic pathway regulation using stress-response promoters[J]. Nature Biotechnology, 2013, 31(11): 1039-1046. |
| 31 | Dhyani R, Shankar K, Bhatt A, et al. Homogentisic acid-based whole-cell biosensor for detection of alkaptonuria disease[J]. Analytical Chemistry, 2021, 93(10): 4521-4527. |
| 32 | Rodrigues A L, Becker J, de Souza Lima A O, et al. Systems metabolic engineering of Escherichia coli for gram scale production of the antitumor drug deoxyviolacein from glycerol[J]. Biotechnology and Bioengineering, 2014, 111(11): 2280-2289. |
| 33 | Guan C, Cui W, Cheng J, et al. Construction and development of an auto-regulatory gene expression system in Bacillus subtilis [J]. Microbial Cell Factories, 2015, 14: 150. |
| 34 | Yin X, Shin H D, Li J H, et al. P gas, a low-pH-induced promoter, as a tool for dynamic control of gene expression for metabolic engineering of Aspergillus niger[J].Applied and Environmental Microbiology, 2017, 83(6): aem03222-16. |
| 35 | Xiong L, Zeng Y, Tang R Q, et al. Condition-specific promoter activities in Saccharomyces cerevisiae[J]. Microbial Cell Factories, 2018, 17(1): 58. |
| 36 | Dabirian Y, Li X, Chen Y, et al. Expanding the dynamic range of a transcription factor-based biosensor in Saccharomyces cerevisiae[J]. ACS Synthetic Biology, 2019, 8(9):1968-1975. |
| 37 | Stella R G, Gertzen C G W, Smits S H J, et al. Biosensor-based growth-coupling and spatial separation as an evolution strategy to improve small molecule production of Corynebacterium glutamicum[J]. Metabolic Engineering, 2021, 68: 162-173. |
| 38 | 邵辉峰, 张斌, 王吕, 等. 钝齿棒杆菌FarR对精氨酸生物合成基因簇转录水平的影响及其与ArgR的关系[J]. 微生物学报, 2014, 54(6): 635-640. |
| Shao H F, Zhang B, Wang L, et al. Effect of FarR on transcriptional levels of arginine biosynthetic genes in Corynebacterium crenatum AS 1.542 and its relationship with ArgR[J]. Acta Microbiologica Sinica, 2014, 54(6): 635-640. | |
| 39 | Sun H, Zhao H, Ang E L. A new biosensor for stilbenes and a cannabinoid enabled by genome mining of a transcriptional regulator[J]. ACS Synthetic Biology, 2020, 9(4): 698-705. |
| 40 | Wachsmuth M, Findeiß S, Weissheimer N, et al. De novo design of a synthetic riboswitch that regulates transcription termination[J]. Nucleic Acids Research, 2013, 41(4): 2541-2551. |
| 41 | You M, Jaffrey S R. Structure and mechanism of RNA mimics of green fluorescent protein[J]. Annual Review of Biophysics, 2015, 44: 187-206. |
| 42 | You M, Litke J L, Jaffrey S R. Imaging metabolite dynamics in living cells using a Spinach-based riboswitch[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(21): E2756-E2765. |
| 43 | Su Y, Hickey S F, Keyser S G, et al. In vitro and in vivo enzyme activity screening via RNA-based fluorescent biosensors for S-adenosyl-l-homocysteine (SAH)[J]. Journal of the American Chemical Society, 2016, 138(22): 7040-7047. |
| 44 | Manna S, Truong J, Hammond M C. Guanidine biosensors enable comparison of cellular turn-on kinetics of riboswitch-based biosensor and reporter[J]. ACS Synthetic Biology, 2021, 10(3): 566-578. |
| 45 | Müller M, Ausländer S, Spinnler A, et al. Designed cell consortia as fragrance-programmable analog-to-digital converters[J]. Nature Chemical Biology, 2017, 13(3): 309-316. |
| 46 | Ganesh I, Ravikumar S, Yoo I K, et al. Construction of malate-sensing Escherichia coli by introduction of a novel chimeric two-component system[J]. Bioprocess and Biosystems Engineering, 2015, 38(4): 797-804. |
| 47 | Kang Z Q, Zhang M M, Gao K Y, et al. An l-2-hydroxyglutarate biosensor based on specific transcriptional regulator LhgR[J]. Nature Communications, 2021, 12: 3619. |
| 48 | Lin J L, Ekas H, Markham K, et al. An enzyme-coupled assay enables rapid protein engineering for geraniol production in yeast[J]. Biochemical Engineering Journal, 2018, 139: 95-100. |
| 49 | Wang B L, Ghaderi A, Zhou H, et al. Microfluidic high-throughput culturing of single cells for selection based on extracellular metabolite production or consumption[J]. Nature Biotechnology, 2014, 32(5): 473-478. |
| 50 | Hanko E K R, Paiva A C, Jonczyk M, et al. A genome-wide approach for identification and characterisation of metabolite-inducible systems[J]. Nature Communications, 2020, 11: 1213. |
| 51 | Mahr R, Frunzke J. Transcription factor-based biosensors in biotechnology: current state and future prospects[J]. Applied Microbiology and Biotechnology, 2016, 100(1): 79-90. |
| 52 | Zhu Y J, Bao Y. Genome-wide mining of MYB transcription factors in the anthocyanin biosynthesis pathway of gossypium hirsutum[J]. Biochemical Genetics, 2021, 59(3): 678-696. |
| 53 | Merulla D, van der Meer J R. Regulatable and modulable background expression control in prokaryotic synthetic circuits by auxiliary repressor binding sites[J]. ACS Synthetic Biology, 2016, 5(1): 36-45. |
| 54 | Siedler S, Schendzielorz G, Binder S, et al. SoxR as a single-cell biosensor for NADPH-consuming enzymes in Escherichia coli[J]. ACS Synthetic Biology, 2014, 3(1): 41-47. |
| 55 | Serganov A, Nudler E. A decade of riboswitches[J]. Cell, 2013, 152(1/2): 17-24. |
| 56 | Rogers J K, Taylor N D, Church G M. Biosensor-based engineering of biosynthetic pathways[J]. Current Opinion in Biotechnology, 2016, 42: 84-91. |
| 57 | Zhang J, Jensen M K, Keasling J D. Development of biosensors and their application in metabolic engineering[J]. Current Opinion in Chemical Biology, 2015, 28: 1-8. |
| 58 | Miyano K, Sudo Y, Yokoyama A, et al. History of the G protein-coupled receptor (GPCR) assays from traditional to a state-of-the-art biosensor assay[J]. Journal of Pharmacological Sciences, 2014, 126(4): 302-309. |
| 59 | Ravikumar S, Baylon M G, Park S J, et al. Engineered microbial biosensors based on bacterial two-component systems as synthetic biotechnology platforms in bioremediation and biorefinery[J]. Microbial Cell Factories, 2017, 16(1): 62. |
| 60 | Ohlendorf R, Vidavski R R, Eldar A, et al. From dusk till dawn: one-plasmid systems for light-regulated gene expression[J]. Journal of Molecular Biology, 2012, 416(4): 534-542. |
| 61 | Peroza E A, Ewald J C, Parakkal G, et al. A genetically encoded Förster resonance energy transfer sensor for monitoring in vivo trehalose-6-phosphate dynamics[J]. Analytical Biochemistry, 2015, 474: 1-7. |
| 62 | Mohsin M, Ahmad A. Genetically-encoded nanosensor for quantitative monitoring of methionine in bacterial and yeast cells [J]. Biosensors and Bioelectronics, 2014, 59: 358-364. |
| 63 | San Martín A, Ceballo S, Baeza-Lehnert F, et al. Imaging mitochondrial flux in single cells with a FRET sensor for pyruvate[J]. PLoS One, 2014, 9(1): e85780. |
| 64 | San Martín A, Ceballo S, Ruminot I, et al. A genetically encoded FRET lactate sensor and its use to detect the Warburg effect in single cancer cells[J]. PLoS One, 2013, 8(2): e57712. |
| 65 | Hessels A M, Chabosseau P, Bakker M H, et al. eZinCh-2: a versatile, genetically encoded FRET sensor for cytosolic and intraorganelle Zn2+ imaging[J]. ACS Chemical Biology, 2015, 10(9): 2126-2134. |
| 66 | Oku M, Hoseki J, Ichiki Y, et al. A fluorescence resonance energy transfer (FRET)-based redox sensor reveals physiological role of thioredoxin in the yeast Saccharomyces cerevisiae[J]. FEBS Letters, 2013, 587(6): 793-798. |
| 67 | Mohsin M, Ahmad A, Iqbal M. FRET-based genetically-encoded sensors for quantitative monitoring of metabolites[J]. Biotechnology Letters, 2015, 37(10): 1919-1928. |
| 68 | Dong H N, Zu X, Zheng P, et al. A rapid enzymatic assay for high-throughput screening of adenosine-producing strains[J]. Microbial Biotechnology, 2015, 8(2): 230-238. |
| 69 | Zhang J, Barajas J F, Burdu M, et al. Development of a transcription factor-based lactam biosensor[J]. ACS Synthetic Biology, 2017, 6(3): 439-445. |
| 70 | Teo W S, Chang M W. Bacterial XylRs and synthetic promoters function as genetically encoded xylose biosensors in Saccharomyces cerevisiae[J]. Biotechnology Journal, 2015, 10(2): 315-322. |
| 71 | Seok J Y, Han Y H, Yang J S, et al. Synthetic biosensor accelerates evolution by rewiring carbon metabolism toward a specific metabolite[J]. Cell Reports, 2021, 36(8): 109589. |
| 72 | Yang P, Wang J, Pang Q X, et al. Pathway optimization and key enzyme evolution of N-acetylneuraminate biosynthesis using an in vivo aptazyme-based biosensor[J]. Metabolic Engineering, 2017, 43: 21-28. |
| 73 | Klauser B, Rehm C, Summerer D, et al. Engineering of ribozyme-based aminoglycoside switches of gene expression by in vivo genetic selection in Saccharomyces cerevisiae[J]. Methods in Enzymology, 2015, 550: 301-320. |
| 74 | Liu Y, Yuan H, Ding D, et al. Establishment of a biosensor-based high-throughput screening platform for tryptophan overproduction[J]. ACS Synthetic Biology, 2021, 10(6):1373-1383. |
| 75 | Lee S W, Oh M K. A synthetic suicide riboswitch for the high-throughput screening of metabolite production in Saccharomyces cerevisiae[J]. Metabolic Engineering, 2015, 28: 143-150. |
| 76 | Lo T M, Chng S H, Teo W S, et al. A two-layer gene circuit for decoupling cell growth from metabolite production[J]. Cell Systems, 2016, 3(2): 133-143. |
| 77 | Yang X Y, Liu J H, Zhang J, et al. Quorum sensing-mediated protein degradation for dynamic metabolic pathway control in Saccharomyces cerevisiae[J]. Metabolic Engineering, 2021, 64: 85-94. |
| 78 | Wu S B, Xue Y T, Yang S J, et al. Combinational quorum sensing devices for dynamic control in cross-feeding cocultivation[J]. Metabolic Engineering, 2021, 67: 186-197. |
| 79 | Rugbjerg P, Sarup-Lytzen K, Nagy M, et al. Synthetic addiction extends the productive life time of engineered Escherichia coli populations[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(10): 2347-2352. |
| 80 | Zhou S H, Yuan S F, Nair P H, et al. Development of a growth coupled and multi-layered dynamic regulation network balancing malonyl-CoA node to enhance (2S)-naringenin biosynthesis in Escherichia coli[J]. Metabolic Engineering, 2021, 67: 41-52. |
| 81 | Flachbart L K, Sokolowsky S, Marienhagen J. Displaced by deceivers: prevention of biosensor cross-talk is pivotal for successful biosensor-based high-throughput screening campaigns[J]. ACS Synthetic Biology, 2019, 8(8): 1847-1857. |
| 82 | Xu P, Li L, Zhang F, et al. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(31): 11299-11304. |
| 83 | Williams T C, Pretorius I S, Paulsen I T. Synthetic evolution of metabolic productivity using biosensors[J]. Trends in Biotechnology, 2016, 34(5): 371-381. |
| 84 | Ko Y S, Kim J W, Lee J A, et al. Tools and strategies of systems metabolic engineering for the development of microbial cell factories for chemical production[J]. Chemical Society Reviews, 2020, 49(14): 4615-4636. |
| 85 | Kaczmarek J A, Prather K L J. Effective use of biosensors for high-throughput library screening for metabolite production[J]. Journal of Industrial Microbiology and Biotechnology, 2021, 8(4):kuab049. |
| 86 | Shen X L, Wang J, Li C Y, et al. Dynamic gene expression engineering as a tool in pathway engineering[J]. Current Opinion in Biotechnology, 2019, 59: 122-129. |
| 87 | Lv Y, Gu Y, Xu J L, et al. Coupling metabolic addiction with negative autoregulation to improve strain stability and pathway yield[J]. Metabolic Engineering, 2020, 61: 79-88. |
| 88 | Wang J, Shen X L, Jain R, et al. Establishing a novel biosynthetic pathway for the production of 3,4-dihydroxybutyric acid from xylose in Escherichia coli[J]. Metabolic Engineering, 2017, 41: 39-45. |
| 89 | Soma Y, Hanai T. Self-induced metabolic state switching by a tunable cell density sensor for microbial isopropanol production [J]. Metabolic Engineering, 2015, 30: 7-15. |
| 90 | Lan E I, Liao J C. ATP drives direct photosynthetic production of 1-butanol in cyanobacteria[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(16): 6018-6023. |
| 91 | Mannan A A, Liu D, Zhang F Z, et al. Fundamental design principles for transcription-factor-based metabolite biosensors[J]. ACS Synthetic Biology, 2017, 6(10): 1851-1859. |
| 92 | Liu Y, Li Q, Zheng P, et al. Developing a high-throughput screening method for threonine overproduction based on an artificial promoter[J]. Microbial Cell Factories, 2015, 14: 121. |
| 93 | Hicks M, Bachmann T T, Wang B J. Synthetic biology enables programmable cell-based biosensors[J]. ChemPhysChem, 2020, 21(2): 132-144. |
| 94 | Ding N N, Yuan Z Q, Zhang X J, et al. Programmable cross-ribosome-binding sites to fine-tune the dynamic range of transcription factor-based biosensor[J]. Nucleic Acids Research, 2020, 48(18): 10602-10613. |
| 95 | Miao Z, Adamiak R W, Antczak M, et al. RNA-Puzzles Round III: 3D RNA structure prediction of five riboswitches and one ribozyme[J]. RNA, 2017, 23(5): 655-672. |
| 96 | Gong S, Wang Y L, Wang Z, et al. Computational methods for modeling aptamers and designing riboswitches[J]. International Journal of Molecular Sciences, 2017, 18(11): 2442. |
| 97 | McKeague M, Wong R S, Smolke C D. Opportunities in the design and application of RNA for gene expression control[J]. Nucleic Acids Research, 2016, 44(7): 2987-2999. |
| 98 | Boyken S E, Benhaim M A, Busch F, et al. De novo design of tunable, pH-driven conformational changes[J]. Science, 2019, 364(6441): 658-664. |
| 99 | Quijano-Rubio A, Yeh H W, Park J, et al. De novo design of modular and tunable protein biosensors[J]. Nature, 2021, 591(7850): 482-487. |
| 100 | Xu P, Wang W Y, Li L Y, et al. Design and kinetic analysis of a hybrid promoter-regulator system for malonyl-CoA sensing in Escherichia coli[J]. ACS Chemical Biology, 2014, 9(2): 451-458. |
| 101 | Fehér T, Libis V, Carbonell P, et al. A sense of balance: experimental investigation and modeling of a malonyl-CoA sensor in Escherichia coli[J]. Frontiers in Bioengineering and Biotechnology, 2015, 3: 46. |
| 102 | Li S J, Si T, Wang M, et al. Development of a synthetic malonyl-CoA sensor in Saccharomyces cerevisiae for intracellular metabolite monitoring and genetic screening[J]. ACS Synthetic Biology, 2015, 4(12): 1308-1315. |
| [1] | Xin LIU, Jun GE, Chun LI. Light-driven microbial hybrid systems improve level of biomanufacturing [J]. CIESC Journal, 2023, 74(1): 330-341. |
| [2] | Xue LIU, Lijuan ZHANG, Guangrong ZHAO. Commensalistic Escherichia coli coculture for biosynthesis of daidzein [J]. CIESC Journal, 2022, 73(9): 4015-4024. |
| [3] | Jingnan WANG, Jian PANG, Lei QIN, Chao GUO, Bo LYU, Chun LI, Chao WANG. Breeding and modification strategies of butenyl-spinosyn high-yield strains [J]. CIESC Journal, 2022, 73(2): 566-576. |
| [4] | Xinhui WANG, Ying WANG, Mingdong YAO, Wenhai XIAO. Research progress of vitamin A biosynthesis [J]. CIESC Journal, 2022, 73(10): 4311-4323. |
| [5] | Wulin ZHOU, Huifang GAO, Yuling WU, Xian ZHANG, Meijuan XU, Taowei YANG, Minglong SHAO, Zhiming RAO. Engineering of Saccharomyces cerevisiae for biosynthesis of campesterol [J]. CIESC Journal, 2021, 72(8): 4314-4324. |
| [6] | WANG Xin, ZHAO Peng, LI Qingyang, TIAN Pingfang. Research advances in semiconductor synthetic biology [J]. CIESC Journal, 2021, 72(5): 2426-2435. |
| [7] | MAO Jinzhu, XIAO Shuling, YANG Zhichun, WANG Xiaoyu, ZHANG Shi, CHEN Junhong, XIE Jisheng, CHEN Fude, HUANG Zinuo, FENG Tianyu, ZHANG Aihui, FANG Baishan. Application of synthetic biology in pesticides residues detection [J]. CIESC Journal, 2021, 72(5): 2413-2425. |
| [8] | Yukun ZHENG, Qing SUN, Zhen CHEN, Huimin YU. Progress for chemicals production via microbial cell factory: selecting several small molecules and macromolecular products as examples [J]. CIESC Journal, 2021, 72(12): 6109-6121. |
| [9] | Zhen ZHANG, Xuecheng ZENG, Lei QIN, Chun LI. Intelligent design of microbial cell factory [J]. CIESC Journal, 2021, 72(12): 6093-6108. |
| [10] | WANG Lian, WU Di, ZHOU Jingwen. Research progress of lignans biosynthesis and their microbial production [J]. CIESC Journal, 2021, 72(1): 320-333. |
| [11] | ZHAO Zhenyao, ZHANG Baocai, LI Feng, SONG Hao. Design and construction of exoelectrogens by synthetic biology [J]. CIESC Journal, 2021, 72(1): 468-482. |
| [12] | WANG Kaifeng, WANG Jinpeng, WEI Ping, JI Xiaojun. Metabolic engineering of Yarrowia lipolytica to produce fatty acids and their derivatives [J]. CIESC Journal, 2021, 72(1): 351-365. |
| [13] | Hutao GAO, Xiaolin SHEN, Xinxiao SUN, Jia WANG, Qipeng YUAN. Metabolic engineering strategies in biosynthesis of amino acids and their derivatives [J]. CIESC Journal, 2020, 71(9): 4058-4070. |
| [14] | Jing XU, Zixuan YOU, Junqi ZHANG, Zheng CHEN, Deguang WU, Feng LI, Hao SONG. Advances in engineering electroactive biofilms by synthetic biology approaches [J]. CIESC Journal, 2020, 71(9): 3950-3962. |
| [15] | Lei QIN, Jie YU, Xiaoyu NING, Wentao SUN, Chun LI. Synthetic biological system construction and green intelligent biological manufacturing [J]. CIESC Journal, 2020, 71(9): 3979-3994. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||