CIESC Journal ›› 2021, Vol. 72 ›› Issue (12): 6298-6310.DOI: 10.11949/0438-1157.20211340
• Biochemical engineering and technology • Previous Articles Next Articles
Nan SU1( ),Yinan WU1(
),Yinan WU1( ),Yinyee TAN1,Lihua JIN2,Chong ZHANG1,Aikawa SHIMPEI3,Hasunuma TOMOHISA3,Kondo AKIHIKO3,Xinhui XING1,4,5(
),Yinyee TAN1,Lihua JIN2,Chong ZHANG1,Aikawa SHIMPEI3,Hasunuma TOMOHISA3,Kondo AKIHIKO3,Xinhui XING1,4,5( )
)
Received:2021-09-16
Revised:2021-11-23
Online:2021-12-22
Published:2021-12-05
Contact:
Xinhui XING
苏楠1( ),吴亦楠1(
),吴亦楠1( ),陈韵亿1,金丽华2,张翀1,Aikawa Shimpei3,Hasunuma Tomohisa3,Kondo Akihiko3,邢新会1,4,5(
),陈韵亿1,金丽华2,张翀1,Aikawa Shimpei3,Hasunuma Tomohisa3,Kondo Akihiko3,邢新会1,4,5( )
)
通讯作者:
邢新会
作者简介:苏楠(1983—),男,博士,基金资助:CLC Number:
Nan SU, Yinan WU, Yinyee TAN, Lihua JIN, Chong ZHANG, Aikawa SHIMPEI, Hasunuma TOMOHISA, Kondo AKIHIKO, Xinhui XING. Comparative omics study of Spirulinaplatensis mutants based on ARTP mutagenesis breeding system[J]. CIESC Journal, 2021, 72(12): 6298-6310.
苏楠, 吴亦楠, 陈韵亿, 金丽华, 张翀, Aikawa Shimpei, Hasunuma Tomohisa, Kondo Akihiko, 邢新会. ARTP诱变钝顶螺旋藻突变体比较组学研究[J]. 化工学报, 2021, 72(12): 6298-6310.
Add to citation manager EndNote|Ris|BibTeX
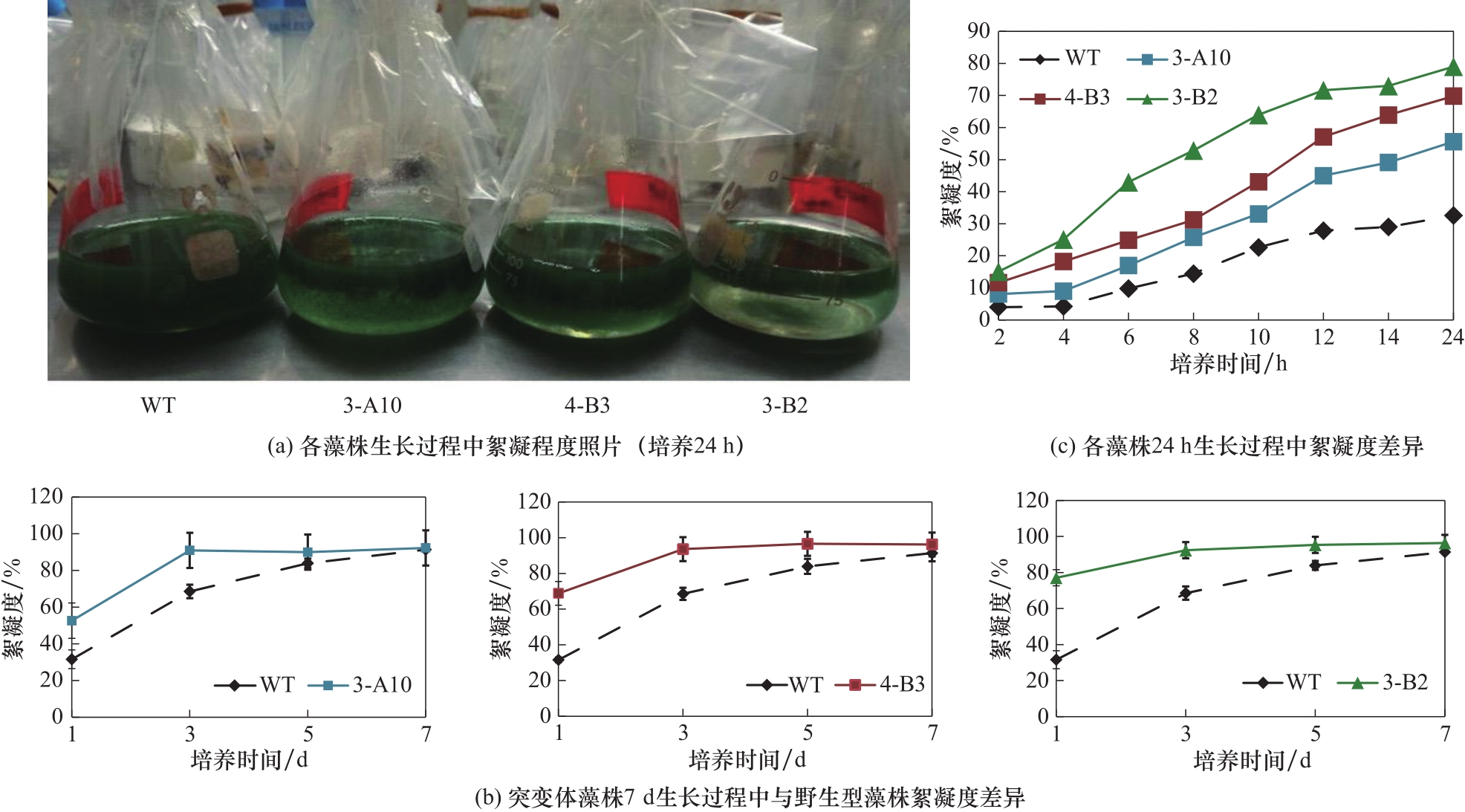
Fig.2 Change of flocculation over culture time. (a) Pictures of S. platensis strain cultures after 24 h incubation in the shaker; (b) The change of flocculation of strain cultures during 7 d of incubation in the shaker; (c) The change of flocculation of strain cultures (harvested after 24 h of incubation in the shaker) after stationary sedimentation in tubes for different time periods
| 聚类分析 | 3-A10 | 3-B2 | 4-B3 |
|---|---|---|---|
| 生物学过程相关 (BP) | 910 | 913 | 910 |
| 细胞组分相关 (CC) | 103 | 101 | 101 |
| 分子功能相关 (MF) | 668 | 672 | 667 |
Table 1 Gene Ontology clustering analysis of mutated strains
| 聚类分析 | 3-A10 | 3-B2 | 4-B3 |
|---|---|---|---|
| 生物学过程相关 (BP) | 910 | 913 | 910 |
| 细胞组分相关 (CC) | 103 | 101 | 101 |
| 分子功能相关 (MF) | 668 | 672 | 667 |
| 序列差异性 | 对照 | 3-A10 | 3-B2 | 4-B3 |
|---|---|---|---|---|
| 单碱基突变数量 | 241873 | 243818 | 241020 | 244320 |
| 过滤野生型基因后的数量 | — | 4063 | 2532 | 4536 |
| 同义单碱基突变 | — | 1040 | 681 | 1229 |
| 错义单碱基突变 | — | 1591 | 1059 | 1823 |
| 进化率 | — | 1.53 | 1.55 | 1.48 |
| 序列插入突变数量 | 345 | 353 | 330 | 362 |
| 过滤野生型基因后的数量 | — | 134 | 101 | 123 |
| 错义插入突变 | — | 82 | 56 | 81 |
Table A1 Statistics on the number of mutations in genome sequencing of Spirulina platensis
| 序列差异性 | 对照 | 3-A10 | 3-B2 | 4-B3 |
|---|---|---|---|---|
| 单碱基突变数量 | 241873 | 243818 | 241020 | 244320 |
| 过滤野生型基因后的数量 | — | 4063 | 2532 | 4536 |
| 同义单碱基突变 | — | 1040 | 681 | 1229 |
| 错义单碱基突变 | — | 1591 | 1059 | 1823 |
| 进化率 | — | 1.53 | 1.55 | 1.48 |
| 序列插入突变数量 | 345 | 353 | 330 | 362 |
| 过滤野生型基因后的数量 | — | 134 | 101 | 123 |
| 错义插入突变 | — | 82 | 56 | 81 |
| 代谢途径 | 突变体 | 代谢途径 | 突变体 | 代谢途径 | 突变体 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 3-A10 | 3-B2 | 4-B3 | 3-A10 | 3-B2 | 4-B3 | 3-A10 | 3-B2 | 4-B3 | |||
| 糖酵解 | 9 | 9 | 9 | 苯丙氨酸代谢 | 2 | 2 | 2 | 乙醛酸、二羧酸代谢 | 4 | 4 | 4 |
| 三羧酸循环 | 2 | 2 | 2 | 色氨酸代谢 | 1 | 1 | 1 | 丙酸代谢 | 1 | 1 | 1 |
| 磷酸戊糖途径 | 7 | 6 | 7 | 苯丙氨酸、酪氨酸、色氨酸合成 | 10 | 10 | 10 | 苯乙烯降解 | 1 | 1 | 1 |
| 果糖、甘露糖代谢 | 5 | 5 | 5 | 叶酸合成 | 8 | 8 | 8 | ||||
| 半乳糖代谢 | 4 | 4 | 4 | 新生毒素生物合成 | 1 | 1 | 1 | 甲烷代谢 | 6 | 6 | 6 |
| 脂肪酸合成 | 3 | 3 | 3 | β-丙氨酸代谢 | 2 | 2 | 2 | 碳固定及光合作用 | 8 | 8 | 8 |
| 氧化磷酸化 | 5 | 5 | 5 | 牛磺酸代谢 | 1 | 1 | 1 | 原核生物碳固定途径 | 6 | 5 | 5 |
| 光合作用 | 2 | 2 | 2 | 有机含硒化合物代谢 | 2 | 2 | 2 | 硫胺素代谢 | 3 | 3 | 3 |
| 嘌呤代谢 | 17 | 17 | 17 | 氨基甲酸代谢 | 2 | 2 | 2 | 核黄素代谢 | 7 | 7 | 7 |
| 丙氨酸、天冬氨酸、谷氨酸代谢 | 5 | 5 | 4 | D-谷氨酰胺、D-谷氨酸代谢 | 1 | 1 | 1 | 维生素B6代谢 | 1 | 1 | 1 |
| 烟酸盐、烟酰胺代谢 | 3 | 3 | 3 | ||||||||
| 四环素合成 | 1 | 1 | 1 | D-丙氨酸代谢 | 1 | 1 | 1 | 泛酸盐及辅酶a生物合成 | 8 | 8 | 8 |
| 黄曲霉毒素合成 | 1 | 1 | 1 | 谷胱甘肽代谢 | 7 | 7 | 7 | ||||
| 甘氨酸、丝氨酸、苏氨酸代谢 | 4 | 4 | 4 | 淀粉、蔗糖代谢 | 7 | 7 | 7 | 生物素代谢 | 1 | 1 | 1 |
| 氨基糖、核苷酸糖代谢 | 6 | 6 | 6 | 叶酸生物合成 | 3 | 3 | 3 | ||||
| 半胱氨酸、蛋氨酸代谢 | 6 | 5 | 6 | 链霉素生物合成 | 5 | 5 | 5 | 阿特拉津降解 | 1 | 1 | 1 |
| 缬氨酸、亮氨酸、异亮氨酸生物合成 | 2 | 2 | 2 | 聚酮糖单元生物合成 | 2 | 2 | 2 | 卟啉、叶绿素代谢 | 15 | 15 | 15 |
| 丁松香和新霉素生物合成 | 2 | 2 | 2 | 萜类化合物生物合成 | 3 | 3 | 3 | ||||
| 赖氨酸生物合成 | 2 | 2 | 2 | ||||||||
| 精氨酸、脯氨酸代谢 | 8 | 8 | 8 | 脂多糖生物合成 | 1 | 1 | 0 | 氮代谢 | 7 | 7 | 7 |
| 组氨酸代谢 | 4 | 4 | 4 | 肽聚糖生物合成 | 3 | 3 | 3 | 糖代谢 | 6 | 6 | 6 |
| 氨基苯甲酸降解 | 2 | 2 | 甘油酯代谢 | 2 | 2 | 2 | 苯丙素生物合成 | 1 | 1 | 1 | |
| 药物代谢相关酶类 | 2 | 2 | 2 | 磷酸肌醇代谢 | 2 | 2 | 2 | 氨酰基-tRNA生物合成 | 6 | 6 | 6 |
| 磷脂酰肌醇信号系统 | 1 | 1 | 1 | 甘油磷脂代谢 | 4 | 4 | 4 | ||||
| 丙酮酸代谢 | 7 | 7 | 7 | 鞘脂类代谢 | 2 | 2 | 2 | 细胞色素P450代谢 | 1 | 1 | 1 |
| 鞘糖脂生物合成 | 1 | 1 | 1 | 鞘糖脂生物合成乳糖 | 1 | 1 | 1 | ||||
Table A2 Metabolic network analysis of mutant genes
| 代谢途径 | 突变体 | 代谢途径 | 突变体 | 代谢途径 | 突变体 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 3-A10 | 3-B2 | 4-B3 | 3-A10 | 3-B2 | 4-B3 | 3-A10 | 3-B2 | 4-B3 | |||
| 糖酵解 | 9 | 9 | 9 | 苯丙氨酸代谢 | 2 | 2 | 2 | 乙醛酸、二羧酸代谢 | 4 | 4 | 4 |
| 三羧酸循环 | 2 | 2 | 2 | 色氨酸代谢 | 1 | 1 | 1 | 丙酸代谢 | 1 | 1 | 1 |
| 磷酸戊糖途径 | 7 | 6 | 7 | 苯丙氨酸、酪氨酸、色氨酸合成 | 10 | 10 | 10 | 苯乙烯降解 | 1 | 1 | 1 |
| 果糖、甘露糖代谢 | 5 | 5 | 5 | 叶酸合成 | 8 | 8 | 8 | ||||
| 半乳糖代谢 | 4 | 4 | 4 | 新生毒素生物合成 | 1 | 1 | 1 | 甲烷代谢 | 6 | 6 | 6 |
| 脂肪酸合成 | 3 | 3 | 3 | β-丙氨酸代谢 | 2 | 2 | 2 | 碳固定及光合作用 | 8 | 8 | 8 |
| 氧化磷酸化 | 5 | 5 | 5 | 牛磺酸代谢 | 1 | 1 | 1 | 原核生物碳固定途径 | 6 | 5 | 5 |
| 光合作用 | 2 | 2 | 2 | 有机含硒化合物代谢 | 2 | 2 | 2 | 硫胺素代谢 | 3 | 3 | 3 |
| 嘌呤代谢 | 17 | 17 | 17 | 氨基甲酸代谢 | 2 | 2 | 2 | 核黄素代谢 | 7 | 7 | 7 |
| 丙氨酸、天冬氨酸、谷氨酸代谢 | 5 | 5 | 4 | D-谷氨酰胺、D-谷氨酸代谢 | 1 | 1 | 1 | 维生素B6代谢 | 1 | 1 | 1 |
| 烟酸盐、烟酰胺代谢 | 3 | 3 | 3 | ||||||||
| 四环素合成 | 1 | 1 | 1 | D-丙氨酸代谢 | 1 | 1 | 1 | 泛酸盐及辅酶a生物合成 | 8 | 8 | 8 |
| 黄曲霉毒素合成 | 1 | 1 | 1 | 谷胱甘肽代谢 | 7 | 7 | 7 | ||||
| 甘氨酸、丝氨酸、苏氨酸代谢 | 4 | 4 | 4 | 淀粉、蔗糖代谢 | 7 | 7 | 7 | 生物素代谢 | 1 | 1 | 1 |
| 氨基糖、核苷酸糖代谢 | 6 | 6 | 6 | 叶酸生物合成 | 3 | 3 | 3 | ||||
| 半胱氨酸、蛋氨酸代谢 | 6 | 5 | 6 | 链霉素生物合成 | 5 | 5 | 5 | 阿特拉津降解 | 1 | 1 | 1 |
| 缬氨酸、亮氨酸、异亮氨酸生物合成 | 2 | 2 | 2 | 聚酮糖单元生物合成 | 2 | 2 | 2 | 卟啉、叶绿素代谢 | 15 | 15 | 15 |
| 丁松香和新霉素生物合成 | 2 | 2 | 2 | 萜类化合物生物合成 | 3 | 3 | 3 | ||||
| 赖氨酸生物合成 | 2 | 2 | 2 | ||||||||
| 精氨酸、脯氨酸代谢 | 8 | 8 | 8 | 脂多糖生物合成 | 1 | 1 | 0 | 氮代谢 | 7 | 7 | 7 |
| 组氨酸代谢 | 4 | 4 | 4 | 肽聚糖生物合成 | 3 | 3 | 3 | 糖代谢 | 6 | 6 | 6 |
| 氨基苯甲酸降解 | 2 | 2 | 甘油酯代谢 | 2 | 2 | 2 | 苯丙素生物合成 | 1 | 1 | 1 | |
| 药物代谢相关酶类 | 2 | 2 | 2 | 磷酸肌醇代谢 | 2 | 2 | 2 | 氨酰基-tRNA生物合成 | 6 | 6 | 6 |
| 磷脂酰肌醇信号系统 | 1 | 1 | 1 | 甘油磷脂代谢 | 4 | 4 | 4 | ||||
| 丙酮酸代谢 | 7 | 7 | 7 | 鞘脂类代谢 | 2 | 2 | 2 | 细胞色素P450代谢 | 1 | 1 | 1 |
| 鞘糖脂生物合成 | 1 | 1 | 1 | 鞘糖脂生物合成乳糖 | 1 | 1 | 1 | ||||
| 1 | Miyamoto K. Renewable Biological Systems for Alternative Sustainable Energy Production[M]. Netherlands: Elsevier Ltd., 1997. |
| 2 | Jagadevan S, Banerjee A, Banerjee C, et al. Recent developments in synthetic biology and metabolic engineering in microalgae towards biofuel production[J]. Biotechnology for Biofuels, 2018, 11: 185. |
| 3 | Casazza A A, Spennati E, Converti A, et al. Production of carbon-based biofuels by pyrolysis of exhausted Arthrospira platensis biomass after protein or lipid recovery[J]. Fuel Processing Technology, 2020, 201: 106336. |
| 4 | Khan M I, Shin J H, Kim J D. The promising future of microalgae: current status, challenges, and optimization of a sustainable and renewable industry for biofuels, feed, and other products[J]. Microbial Cell Factories, 2018, 17(1): 36. |
| 5 | Liestianty D, Rodianawati I, Arfah R A, et al. Nutritional analysis of Spirulina sp. to promote as superfood candidate[J]. IOP Conference Series: Materials Science and Engineering, 2019, 509: 012031. |
| 6 | Nasirian F, Dadkhah M, Moradi-Kor N, et al. Effects of Spirulina platensis microalgae on antioxidant and anti-inflammatory factors in diabetic rats[J]. Diabetes, Metabolic Syndrome and Obesity: Targets and Therapy, 2018, 11: 375-380. |
| 7 | Aydi S S, Aydi S, Ben Abdallah Kolsi R, et al. CO2 enrichment: enhancing antioxidant, antibacterial and anticancer activities in Arthrospira platensis[J]. Food Bioscience, 2020, 35: 100575. |
| 8 | Saranraj P, Sivasakthi S. Spirulina platensis-food for future: a review[J]. Asian J. Pharm. Sci. Technol., 2014, 4(1): 26-33. |
| 9 | Mukhopadhyay C D. Engineering spirulina for enhanced medicinal application[M]//Algal Biorefinery: An Integrated Approach. Cham: Springer International Publishing, 2015: 235-252. |
| 10 | Saini D K, Chakdar H, Pabbi S, et al. Enhancing production of microalgal biopigments through metabolic and genetic engineering[J]. Critical Reviews in Food Science and Nutrition, 2020, 60(3): 391-405. |
| 11 | Ragusa I, Nardone G N, Zanatta S, et al. Spirulina for skin care: a bright blue future[J]. Cosmetics, 2021, 8(1): 7. |
| 12 | Rempel A, de Souza Sossella F, Margarites A C, et al. Bioethanol from Spirulina platensis biomass and the use of residuals to produce biomethane: an energy efficient approach[J]. Bioresource Technology, 2019, 288: 121588. |
| 13 | Kawata Y, Yano S, Kojima H, et al. Transformation of Spirulina platensis strain C1 (Arthrospira sp. PCC9438) with Tn5 transposase-transposon DNA-cation liposome complex[J]. Marine Biotechnology, 2004, 6(4): 355-363. |
| 14 | Dehghani J, Adibkia K, Movafeghi A, et al. Stable transformation of Spirulina (Arthrospira) platensis: a promising microalga for production of edible vaccines[J]. Applied Microbiology and Biotechnology, 2018, 102(21): 9267-9278. |
| 15 | Jeamton W, Dulsawat S, Tanticharoen M, et al. Overcoming intrinsic restriction enzyme barriers enhances transformation efficiency in Arthrospira platensis C1[J]. Plant and Cell Physiology, 2017, 58(4): 822-830. |
| 16 | Cheevadhanarak S, Paithoonrangsarid K, Prommeenate P, et al. Draft genome sequence of Arthrospira platensis C1 (PCC9438)[J]. Standards in Genomic Sciences, 2012, 6(1): 43-53. |
| 17 | 殷春涛, 胡鸿钧, 李夜光, 等. 中温螺旋藻新品系的选育研究[J]. 武汉植物学研究, 1997, 15(3): 250-254. |
| Yin C T, Hu H J, Li Y G, et al. Studies on middle temperature strains selection of Spirulina platensis[J]. Journal of Wuhan Botanical Research, 1997, 15(3): 250-254. | |
| 18 | 李建宏, 郑卫, 倪霞, 等. 两株钝顶螺旋藻紫外诱变株的特征[J]. 水生生物学报, 2001, 25(5): 486-490. |
| Li J H, Zheng W, Ni X, et al. Characteristics of two Spirulina platensis mutants induced by ultraviolet[J]. Acta Hydrobiologica Sinica, 2001, 25(5): 486-490. | |
| 19 | Fang M, Jin L, Zhang C, et al. Rapid mutation of Spirulina platensis by a new mutagenesis system of atmospheric and room temperature plasmas (ARTP) and generation of a mutant library with diverse phenotypes[J]. PLoS One, 2013, 8(10): e77046. |
| 20 | Shirnalli G G, Kaushik M S, Kumar A, et al. Isolation and characterization of high protein and phycocyanin producing mutants of Arthrospira platensis[J]. Journal of Basic Microbiology, 2018, 58(2): 162-171. |
| 21 | Cheng J, Lu H X, He X, et al. Mutation of Spirulina sp. by nuclear irradiation to improve growth rate under 15% carbon dioxide in flue gas[J]. Bioresource Technology, 2017, 238: 650-656. |
| 22 | Zhu Y X, Cheng J, Zhang Z, et al. Mutation of Arthrospira platensis by gamma irradiation to promote phenol tolerance and CO2 fixation for coal-chemical flue gas reduction[J]. Journal of CO2 Utilization, 2020, 38: 252-261. |
| 23 | An J, Gao F L, Ma Q Y, et al. Screening for enhanced astaxanthin accumulation among Spirulina platensis mutants generated by atmospheric and room temperature plasmas[J]. Algal Research, 2017, 25: 464-472. |
| 24 | Fujisawa T, Narikawa R, Okamoto S, et al. Genomic structure of an economically important cyanobacterium, Arthrospira (Spirulina) platensis NIES-39[J]. DNA Research, 2010, 17(2): 85-103. |
| 25 | Cheng J, Lu H X, Li K, et al. Enhancing growth-relevant metabolic pathways of Arthrospira platensis (CYA-1) with gamma irradiation from 60Co[J]. RSC Advances, 2018, 8(30): 16824-16833. |
| 26 | Kurdrid P, Senachak J, Sirijuntarut M, et al. Comparative analysis of the Spirulina platensis subcellular proteome in response to low- and high-temperature stresses: uncovering cross-talk of signaling components[J]. Proteome Sci., 2011, 9: 39. |
| 27 | Depraetere O, Deschoenmaeker F, Badri H, et al. Trade-off between growth and carbohydrate accumulation in nutrient-limited Arthrospira sp. PCC 8005 studied by integrating transcriptomic and proteomic approaches[J]. PLoS One, 2015, 10(7): e0132461. |
| 28 | Kumaresan V, Nizam F, Ravichandran G, et al. Transcriptome changes of blue-green algae, Arthrospira sp. in response to sulfate stress[J]. Algal Research, 2017, 23: 96-103. |
| 29 | Ismaiel M M S, Piercey-Normore M D, Rampitsch C. Proteomic analyses of the cyanobacterium Arthrospira (Spirulina) platensis under iron and salinity stress[J]. Environmental and Experimental Botany, 2018, 147: 63-74. |
| 30 | Tan Y, Fang M Y, Jin L H, et al. Culture characteristics of the atmospheric and room temperature plasma-mutated Spirulina platensis mutants in CO2 aeration culture system for biomass production[J]. Journal of Bioscience and Bioengineering, 2015, 120(4): 438-443. |
| 31 | Vonshak A. Spirulina platensis (Arthrospira): Physiology, Cell Biology and Biotechnology[M]. London: CRC Press, 1997. |
| [1] | Tongpeng LU, Xiaolin PAN, Hongfei WU, Yu LI, Haiyan YU. Effect of organic flocculant on settling performance of iron-bearing minerals and its adsorption mechanism [J]. CIESC Journal, 2022, 73(9): 4122-4132. |
| [2] | Caifeng LI, Xiao WANG, Gangjian LI, Junzhang LIN, Weidong WANG, Qinglin SHU, Yanbin CAO, Meng XIAO. Synergistic relationship between hydrocarbon degrading and emulsifying strain SL-1 and endogenous bacteria during oil displacement [J]. CIESC Journal, 2022, 73(9): 4095-4102. |
| [3] | Mai ZHANG, Yao TIAN, Zhiqi GUO, Ye WANG, Guangjin DOU, Hao SONG. Design and optimization of photocatalysis-biological hybrid system for green synthesis of fuels and chemicals [J]. CIESC Journal, 2022, 73(7): 2774-2789. |
| [4] | Jingnan WANG, Jian PANG, Lei QIN, Chao GUO, Bo LYU, Chun LI, Chao WANG. Breeding and modification strategies of butenyl-spinosyn high-yield strains [J]. CIESC Journal, 2022, 73(2): 566-576. |
| [5] | Ming HUANG, Liang ZHU, Zixia DING, Yiting MAO, Zhongqing MA. Synergistic interactions of biomass three-component and low-density polyethylene during co-catalytic fast pyrolysis for the production of light aromatics [J]. CIESC Journal, 2022, 73(2): 699-711. |
| [6] | Xiaosong HOU, Chenxing LIU, Ailing REN, Bin GUO, Yuanming GUO. Study on purification of toluene waste gas by ultrasonic atomization/surfactants-enhanced absorption coupled with biological scrubbing [J]. CIESC Journal, 2022, 73(10): 4692-4706. |
| [7] | Haibo LIU, Nan WANG, Hongzhou LIU, Tiezhu CHEN, Jianchang LI. Effects of voltage perturbation on the activities of microorganisms and key enzymes in EAD metabolic flux [J]. CIESC Journal, 2022, 73(10): 4603-4612. |
| [8] | Wei SONG, Jinhui WANG, Guipeng HU, Xiulai CHEN, Liming LIU, Jing WU. Cascade catalysis for the synthesis of (R)-β-tyrosine [J]. CIESC Journal, 2022, 73(1): 352-361. |
| [9] | CHEN Tingting, HAN Kaixin, CHEN Cuixue, LING Xueping, SHEN Liang, LU Yinghua. Study of iron-reducing bacteria Shewanellaxiamenensis BC01 under organic solvents stress [J]. CIESC Journal, 2021, 72(7): 3747-3756. |
| [10] | MAO Jinzhu, XIAO Shuling, YANG Zhichun, WANG Xiaoyu, ZHANG Shi, CHEN Junhong, XIE Jisheng, CHEN Fude, HUANG Zinuo, FENG Tianyu, ZHANG Aihui, FANG Baishan. Application of synthetic biology in pesticides residues detection [J]. CIESC Journal, 2021, 72(5): 2413-2425. |
| [11] | ZHAO Zhenyao, ZHANG Baocai, LI Feng, SONG Hao. Design and construction of exoelectrogens by synthetic biology [J]. CIESC Journal, 2021, 72(1): 468-482. |
| [12] | WANG Kaifeng, WANG Jinpeng, WEI Ping, JI Xiaojun. Metabolic engineering of Yarrowia lipolytica to produce fatty acids and their derivatives [J]. CIESC Journal, 2021, 72(1): 351-365. |
| [13] | Lei QIN, Jie YU, Xiaoyu NING, Wentao SUN, Chun LI. Synthetic biological system construction and green intelligent biological manufacturing [J]. CIESC Journal, 2020, 71(9): 3979-3994. |
| [14] | Yan JIANG, Zhe ZHANG. Interaction of VOCs with different hydrophilic properties in biotrickling filters [J]. CIESC Journal, 2020, 71(7): 2973-2982. |
| [15] | Hongwei JIN,Dandan ZHAI,Xin WANG,Shuang ZHAO,Xiangyang MENG,Yueying HE,Yang SHEN,Ming HUI. Effect of graphene/polyaniline modified anode on performance of microbial fuel cell [J]. CIESC Journal, 2019, 70(6): 2343-2350. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||