化工学报 ›› 2020, Vol. 71 ›› Issue (7): 2945-2955.DOI: 10.11949/0438-1157.20200070
封悦洋1,2,3( ),王颖2,3,姚明东1,2,3(
),王颖2,3,姚明东1,2,3( ),肖文海2,3,丁明珠2,3
),肖文海2,3,丁明珠2,3
收稿日期:2020-01-16
修回日期:2020-04-06
出版日期:2020-07-05
发布日期:2020-07-05
通讯作者:
姚明东
作者简介:封悦洋(1995—),女,硕士研究生,基金资助:
Yueyang FENG1,2,3( ),Ying WANG2,3,Mingdong YAO1,2,3(
),Ying WANG2,3,Mingdong YAO1,2,3( ),Wenhai XIAO2,3,Mingzhu DING2,3
),Wenhai XIAO2,3,Mingzhu DING2,3
Received:2020-01-16
Revised:2020-04-06
Online:2020-07-05
Published:2020-07-05
Contact:
Mingdong YAO
摘要:
糖苷类天然产物是植物中次级代谢产物的主要存在形式,具有重要的生物活性。通过糖基化修饰可以改变其水溶性、稳定性,产生特殊的生物活性和功能,因此易于商品化应用,具有重要的药用价值和工业价值,尤其在抗癌药物和日化用品中有着广泛应用。近年来糖苷类产物的生物合成也取得了重大的进展。以重要的糖苷化合物槲皮素为例介绍了糖苷化合物的生物合成,从糖基转移酶和前体供应等角度阐述了生物高产糖苷类化合物的工程策略,为利用合成生物学技术获得植物糖苷的高产菌株提供了技术参考。
中图分类号:
封悦洋, 王颖, 姚明东, 肖文海, 丁明珠. 生物合成槲皮素糖苷类衍生物的研究进展[J]. 化工学报, 2020, 71(7): 2945-2955.
Yueyang FENG, Ying WANG, Mingdong YAO, Wenhai XIAO, Mingzhu DING. Advances in biosynthesis of quercetin glycoside derivatives[J]. CIESC Journal, 2020, 71(7): 2945-2955.
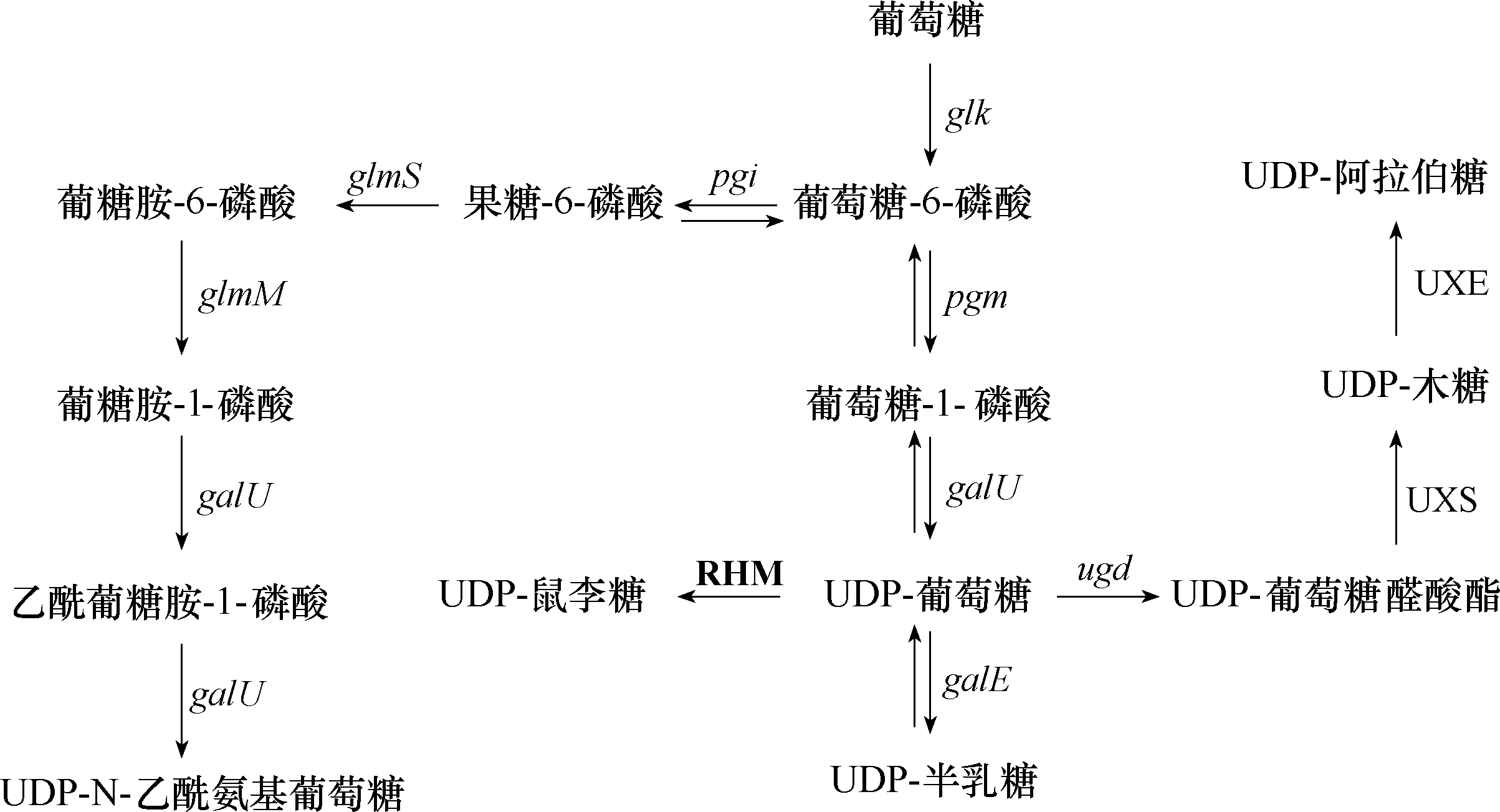
图3 UDP-sugars生物合成途径粗体大写文字表示存在于植物中,其他存在于细菌中。glk—葡萄糖激酶编码基因;pgm—磷酸变位酶编码基因;pgi—磷酸葡糖异构酶编码基因;galU—UTP-葡萄糖-1-磷酸尿苷转移酶编码基因;galE—UDP-葡萄糖-4-表异构酶编码基因;ugd—UDP-葡萄糖-6-脱氢酶编码基因;glmS—果糖-6-磷酸氨基转移酶编码基因;glmM—磷酸葡萄糖胺突变酶编码基因; UXE—UDP-木糖-4-表异构酶;UXS—UDP-木糖合酶;RHM—鼠李糖合成酶
Fig.3 UDP-sugars biosynthesis pathway
| 糖苷 | 糖供体 | 糖基转移酶 | UDP-sugars生物合成相关基因 | 产量/(mg/L) | 文献 |
|---|---|---|---|---|---|
| quercetin-4'-O-glucoside | UDPG | AtUGT74F1 | — | 0.19 | [ |
| quercetin-3'-O-glucoside | UDPG | AtUGT71C1 | — | 8 | [ |
| quercetin-3-O-glucoside | UDPG | AtUGT73B3 | — | 99.3 | [ |
| quercetin-3-O-glucoside | UDPG | AtUGT73B3 | Δpgi | 3900 | [ |
| quercetin-7-O-glucoside | UDPG | AtUGT76E12 | — | 4.74 | [ |
| quercetin-7-O-glucoside | UDPG | AtUGT84B1 | Δpgi | 95 | [ |
| quercetin-3-O-rhamnoside | UDP-Rha | AtUGT78D1 | RHM2 | 150 | [ |
| quercetin-3-O-rhamnoside | UDP-Rha | AtGT | BaSP, MUM4, Δpgm, ΔushA, Δagp, ΔgalETKM | 1120 | [ |
| quercetin-3-O-galactoside | UDP-Gal | PhF3GT | BaSP, galE, Δpgm, ΔushA, Δagp, ΔgalETKM | 940 | [ |
| quercetin-3-O-galactoside | UDP-Gal | PhUGT | GmSUS-galE | 2134 | [ |
| quercetin-3-O-xyloside | UDP-Xyl | AtGT3 | naf44530-galU, calS8, calS9, Δpgi, Δzwf, ΔushA | 23.78 | [ |
| quercetin-3-O-xyloside | UDP-Xyl | AtUGT78D3(E380Q) | ecugd, AtUXS, ΔarnA | 150 | [ |
| quercetin-3-O-arabinoside | UDP-Ara | AtUGT78D3 | ecugd, AtUXS, OsUXE, ΔarnA | 158 | [ |
| quercetin-3-O-glucuronide | UDP-GlcA | VvUGT | ecugd, ΔarnA | 687 | [ |
| quercetin-3-O-N-acetylglucosamine | UDP-GlcNAc | AtUGT78D2 | galU | 380 | [ |
| quercetin-3,7-di-O-glucoside | UDPG | AtUGT76E12 | — | 0.71 | [ |
| quercetin-7,3'-di-O-glucoside | UDPG | AtUGT71C1 | — | 10.9 | [ |
| quercetin-3-O-glucoside-7-O-rhamnoside | UDPG UDP-Rha | AtUGT78D2 AtUGT89C1 | — | 67 | [ |
| quercetin-3-O-glucosyl(1→2)xyloside | UDPG UDP-Xyl | AtUGT78D2 AtUGT79B1 | ecugd, AtUXS | 65 | [ |
| quercetin-3-O-glucosyl(1→6)rhamnoside | UDPG UDP-Rha | BcUGT1 GmFg2 | AtRHM2 | 119.8 | [ |
| quercetin-3-O-glucuronic acid-7-O-rhamnoside | UDP-GlcA UDP-Rha | VvUGT AtUGT89C1 | ecugd, AtRHM2 | 44.8 | [ |
| quercetin-3-O-arabinosE-7-O- rhamnoside | UDP-Ara UDP-Rha | AtUGT78D3 AtUGT89C1 | OsUXE, AtUXS, ecugd, AtRHM | 45.1 | [ |
表1 槲皮素糖苷在大肠杆菌中的生物合成
Table 1 Biosynthesis of quercetin glycosides in E. coli
| 糖苷 | 糖供体 | 糖基转移酶 | UDP-sugars生物合成相关基因 | 产量/(mg/L) | 文献 |
|---|---|---|---|---|---|
| quercetin-4'-O-glucoside | UDPG | AtUGT74F1 | — | 0.19 | [ |
| quercetin-3'-O-glucoside | UDPG | AtUGT71C1 | — | 8 | [ |
| quercetin-3-O-glucoside | UDPG | AtUGT73B3 | — | 99.3 | [ |
| quercetin-3-O-glucoside | UDPG | AtUGT73B3 | Δpgi | 3900 | [ |
| quercetin-7-O-glucoside | UDPG | AtUGT76E12 | — | 4.74 | [ |
| quercetin-7-O-glucoside | UDPG | AtUGT84B1 | Δpgi | 95 | [ |
| quercetin-3-O-rhamnoside | UDP-Rha | AtUGT78D1 | RHM2 | 150 | [ |
| quercetin-3-O-rhamnoside | UDP-Rha | AtGT | BaSP, MUM4, Δpgm, ΔushA, Δagp, ΔgalETKM | 1120 | [ |
| quercetin-3-O-galactoside | UDP-Gal | PhF3GT | BaSP, galE, Δpgm, ΔushA, Δagp, ΔgalETKM | 940 | [ |
| quercetin-3-O-galactoside | UDP-Gal | PhUGT | GmSUS-galE | 2134 | [ |
| quercetin-3-O-xyloside | UDP-Xyl | AtGT3 | naf44530-galU, calS8, calS9, Δpgi, Δzwf, ΔushA | 23.78 | [ |
| quercetin-3-O-xyloside | UDP-Xyl | AtUGT78D3(E380Q) | ecugd, AtUXS, ΔarnA | 150 | [ |
| quercetin-3-O-arabinoside | UDP-Ara | AtUGT78D3 | ecugd, AtUXS, OsUXE, ΔarnA | 158 | [ |
| quercetin-3-O-glucuronide | UDP-GlcA | VvUGT | ecugd, ΔarnA | 687 | [ |
| quercetin-3-O-N-acetylglucosamine | UDP-GlcNAc | AtUGT78D2 | galU | 380 | [ |
| quercetin-3,7-di-O-glucoside | UDPG | AtUGT76E12 | — | 0.71 | [ |
| quercetin-7,3'-di-O-glucoside | UDPG | AtUGT71C1 | — | 10.9 | [ |
| quercetin-3-O-glucoside-7-O-rhamnoside | UDPG UDP-Rha | AtUGT78D2 AtUGT89C1 | — | 67 | [ |
| quercetin-3-O-glucosyl(1→2)xyloside | UDPG UDP-Xyl | AtUGT78D2 AtUGT79B1 | ecugd, AtUXS | 65 | [ |
| quercetin-3-O-glucosyl(1→6)rhamnoside | UDPG UDP-Rha | BcUGT1 GmFg2 | AtRHM2 | 119.8 | [ |
| quercetin-3-O-glucuronic acid-7-O-rhamnoside | UDP-GlcA UDP-Rha | VvUGT AtUGT89C1 | ecugd, AtRHM2 | 44.8 | [ |
| quercetin-3-O-arabinosE-7-O- rhamnoside | UDP-Ara UDP-Rha | AtUGT78D3 AtUGT89C1 | OsUXE, AtUXS, ecugd, AtRHM | 45.1 | [ |

图5 大肠杆菌中UDP-sugars内源合成路径优化实线路径代表大肠杆菌内源路径,虚线路径代表外源路径,斜线代表敲除。naf44530—皮诺卡氏菌葡萄糖磷酸变位酶编码基因;zwf—6-磷酸葡萄糖-1-脱氢酶编码基因;ushA—UDPG水解酶编码基因;calS8—UDP-葡萄糖脱氢酶编码基因;calS9—UDP-葡萄糖醛酸脱氢酶编码基因;VvUGT,AtGT3—糖基转移酶;arnA—UDP-葡萄糖醛酸C-4 -脱羧酶编码基因;glk—葡萄糖激酶编码基因;pgm—磷酸变位酶编码基因;pgi—磷酸葡糖异构酶编码基因;galU—UTP-葡萄糖-1-磷酸尿苷转移酶编码基因;ugd—UDP-葡萄糖-6-脱氢酶编码基因
Fig.5 Optimization of UDP-sugars endogenous synthesis pathway in E. coli

图6 大肠杆菌中UDP-sugars合成路径的正交化设计实线路径代表大肠杆菌内源路径,虚线路径代表外源路径,斜线代表敲除。GmSUS—大豆蔗糖合酶;PhUGT,AtGt—糖基转移酶;BaSP—蔗糖磷酸化酶;MUM4—鼠李糖合成酶;galE—UDP-葡萄糖-4-表异构酶编码基因;ugpA—尿苷转移酶编码基因;agp—葡萄糖-1-磷酸酶编码基因;ushA—UDPG水解酶编码基因;galETKM—半乳糖操纵子编码基因;pgm—磷酸变位酶编码基因
Fig.6 Orthogonal design of UDP-sugars synthesis pathway in E. coli
| 1 | Kren V, Martinkova L. Glycosides in medicine: “The role of glycosidic residue in biological activity”[J]. Curr. Med. Chem., 2001, 8(11): 1303-1328. |
| 2 | Jakeman D L, Sadeghi-Khomami A. A β-(1, 2)-glycosynthase and an attempted selection method for the directed evolution of glycosynthases[J]. Biochemistry, 2011, 50(47): 10359-10366. |
| 3 | Liu X, Cheng J, Zhang G, et al. Engineering yeast for the production of breviscapine by genomic analysis and synthetic biology approaches[J]. Nat. Commun., 2018, 9(1): 448. |
| 4 | Shen X, Wang J, Wang J, et al. High-level de novo biosynthesis of arbutin in engineered Escherichia coli[J]. Metab. Eng., 2017, 42: 52-58. |
| 5 | Stahlhut S G, Siedler S, Malla S, et al. Assembly of a novel biosynthetic pathway for production of the plant flavonoid fisetin in Escherichia coli[J]. Metab. Eng., 2015, 31: 84-93. |
| 6 | 文中行, 胡占兴, 袁洁, 等. 槲皮素糖苷类化合物的合成及其对α-葡萄糖苷酶的抑制活性[J]. 山地农业生物学报, 2016, 35(3): 18-24. |
| Wen Z H, Hu Z X, Yuan J, et al. Synthesis of quercetin glycosides and their inhibitory activities to α-glucosidase[J]. Journal of Mountain Agriculture and Biology, 2016, 35(3): 18-24. | |
| 7 | 叶林虎, 闫明珠, 孔令提, 等. 槲皮素及其糖苷类化合物对P450酶活性的体外抑制作用[J]. 中国药学杂志, 2014, 49(12): 1051-1055. |
| Ye L H, Yan M Z, Kong L T, et al. In vitro inhibition of quercetin and its glycosides on P450 enzyme activities[J]. Chin. Pharmacol. J., 2014, 49(12): 1051-1055. | |
| 8 | Lu J, Li J, Wang S, et al. Advances in ginsenoside biosynthesis and metabolic regulation[J]. Biotechnol. Appl. Biochem., 2018, 65(4): 514-522. |
| 9 | Fraser-Reid B, López J C, Gammon D W, et al. Other methods for glycoside synthesis[M]//Demchenko A V. Handbook of Chemical Glycosylation: Advances in Stereoselectivity and Therapeutic Relevance. Weinheim,Germany: Wiley-VCH Verlag GmbH & Co. KGaA, 2008. |
| 10 | Chung D, Kim S Y, Ahn J H. Production of three phenylethanoids, tyrosol, hydroxytyrosol, and salidroside, using plant genes expressing in Escherichia coli[J]. Sci. Rep., 2017, 7(1): 2578. |
| 11 | Lin Y, Sun X, Yuan Q, et al. Combinatorial biosynthesis of plant-specific coumarins in bacteria[J]. Metab. Eng., 2013, 18: 69-77. |
| 12 | Zhuang Y, Yang G, Chen X, et al. Biosynthesis of plant-derived ginsenoside Rh2 in yeast via repurposing a key promiscuous microbial enzyme[J]. Metab. Eng., 2017, 42: 25-32. |
| 13 | Hansen E H, Moller B L, Kock G R, et al. De novo biosynthesis of vanillin in fission yeast (Schizosaccharomyces pombe) and baker s yeast (Saccharomyces cerevisiae)[J]. Appl. Environ. Microbiol., 2009, 75: 2765-2774 |
| 14 | Leonard E, Yan Y, Fowler Z L, et al. Strain improvement of recombinant Escherichia coli for efficient production of plant flavonoids[J]. Mol. Pharm., 2008, 5: 257-265. |
| 15 | Wang X. Structure, mechanism and engineering of plant natural product glycosyltransferases[J]. FEBS Lett., 2009, 583(20): 3303-3309. |
| 16 | Huang F C, Hinkelmann J, Hermenau A. Enhanced production of β-glucosides by in-situ UDP-glucose regeneration[J]. J. Biotechnol., 2016, 224: 35-44. |
| 17 | de Bruyn F, Maertens J, Beauprez J, et al. Biotechnological advances in UDP-sugar based glycosylation of small molecules[J]. Biotechnol. Adv., 2015, 33(2): 288-302. |
| 18 | Lim E K, Ashford D A, Hou B, et al. Arabidopsis glycosyltransferases as biocatalysts in fermentation for regioselective synthesis of diverse quercetin glucosides[J]. Biotechnol. Bioeng., 2004, 87(5): 623-631. |
| 19 | Willits M G, Giovanni M, Prata R T N, et al. Bio-fermentation of modified flavonoids: an example of in vivo diversification of secondary metabolites[J]. Phytochemistry, 2004, 65(1): 31-41. |
| 20 | Kashyap D, Mittal S, Sak K, et al. Molecular mechanisms of action of quercetin in cancer: recent advances[J]. Tumor Biol., 2016, 37(10): 12927-12939. |
| 21 | 闫淑霞, 李鲜, 孙崇德, 等. 槲皮素及其糖苷衍生物降糖降脂活性研究进展[J]. 中国中药杂志, 2015, 40(23): 47-54. |
| Yan S X, Li X, Sun C D, et al. Hypoglycemic and hypolipidemic effects of quercetin and its glycosides[J]. Chin. J. Chin. Mater. Med., 2015, 40(23): 47-54. | |
| 22 | Heo K T, Kang S Y, Hong Y S. De novo biosynthesis of pterostilbene in an Escherichia coli strain using a new resveratrol O-methyltransferase from Arabidopsis[J]. Microb. Cell. Fact., 2017, 16(1): 30. |
| 23 | Reuben S, Rai A, Pillai B V S, et al. A bacterial quercetin oxidoreductase QuoA-mediated perturbation in the phenylpropanoid metabolic network increases lignification with a concomitant decrease in phenolamides in Arabidopsis[J]. J. Exp. Bot., 2013, 64(16): 5183-5194. |
| 24 | Wang Y, Chen S, Yu O. Metabolic engineering of flavonoids in plants and microorganisms[J]. Appl. Microbiol. Biotechnol., 2011, 91: 949-956. |
| 25 | Crozier A, Jaganath I B, Clifford M N. Dietary phenolics: chemistry, bioavailability and effects on health[J].Nat. Prod. Rep., 2009, 26: 1001-1043. |
| 26 | Ververidis F. Metabolic engineering of the complete pathway leading to heterologous biosynthesis of various flavonoids and stilbenoids in Saccharomyces cerevisiae[J]. Metab. Eng., 2009, 11(6): 355-366. |
| 27 | Yan Y, Koffas M A G, Leonard E. Functional expression of a P450 flavonoid hydroxylase for the biosynthesis of plant-specific hydroxylated flavonols in Escherichia coli[J]. Metab. Eng., 2006, 8(2): 172-181. |
| 28 | Marín L, Gutiérrez-del-Río I, Entrialgo-Cadierno R, et al. De novo biosynthesis of myricetin, kaempferol and quercetin in Streptomyces albus and Streptomyces coelicolor[J]. PLoS One, 2018, 13(11): e0207278. |
| 29 | Kallscheuer N, Vogt M, Bott M, et al. Functional expression of plant-derived O-methyltransferase, flavanone 3-hydroxylase, and flavonol synthase in Corynebacterium glutamicum for production of pterostilbene, kaempferol, and quercetin[J]. J. Biotechnol., 2017, 258: 190-196. |
| 30 | Gutierrez A, Grunau A, Paine M, et al. Electron transfer in human cytochrome P450 reductase[J]. Biochem. Soc. Trans., 2003, 31(3): 497-501. |
| 31 | Rodriguez A, Strucko T, Stahlhut S G, et al. Metabolic engineering of yeast for fermentative production of flavonoids[J]. Bioresource Technology, 2017, 245(Pt B): 1645-1654. |
| 32 | Verstrepen K J, Iserentant D, Malcorps P, et al. Glucose and sucrose: hazardous fast-food for industrial yeast?[J]. Trends Biotechnol., 2004, 22(10): 531-537. |
| 33 | Liu X, Li X, Jiang J, et al. Convergent engineering of syntrophic Escherichia coli coculture for efficient production of glycosides[J]. Metab. Eng., 2018, 47: 243-253. |
| 34 | Rosenberger A F N, Hangelmann L, Hofinger A, et al. UDP-xylose and UDP-galactose synthesis in Trichomonas vaginalis[J]. Mol. Biochem. Parasit., 2012, 181(1): 53-56. |
| 35 | Oka T, Jigami Y. Reconstruction of de novo pathway for synthesis of UDP-glucuronic acid and UDP-xylose from intrinsic UDP-glucose in Saccharomyces cerevisiae[J]. FEBS J., 2006, 273(12): 2645-2657. |
| 36 | Kim S Y, Lee H R, Park K S, et al. Metabolic engineering of Escherichia coli for the biosynthesis of flavonoid-O-glucuronides and flavonoid-O-galactoside[J]. Appl. Microbiol. Biotechnol., 2015, 99(5): 2233-2242. |
| 37 | Gu X, Lee S G, Bar-Peled M. Biosynthesis of UDP-xylose and UDP-arabinose in Sinorhizobium meliloti 1021: first characterization of a bacterial UDP-xylose synthase, and UDP-xylose 4-epimerase[J]. Microbiology, 2011, 157(1): 260-269. |
| 38 | Oikari S, Kettunen T, Tiainen S, et al. UDP-sugar accumulation drives hyaluronan synthesis in breast cancer[J]. Matrix Biol., 2018, 67: 63-74. |
| 39 | Dai X L, Zhao G F, Jiao T M, et al. Involvement of three CsRHM genes from Camellia sinensis in UDP-rhamnose biosynthesis[J]. J. Agric. Food Chem., 2018, 66(27): 7139-7149. |
| 40 | Tiwari P, Sangwan R S, Sangwan N S. Plant secondary metabolism linked glycosyltransferases: an update on expanding knowledge and scopes[J]. Biotechnol. Adv., 2016, 34(5): 714-739. |
| 41 | Meech R, Hu D G, McKinnon R A, et al. The UDP-glycosyltransferase (UGT) superfamily: new members, new functions, and novel paradigms[J]. Physiol. Rev., 2019, 99(2): 1153-1222. |
| 42 | Paquette S M, Jensen K, Bak S. A web-based resource for the Arabidopsis P450, cytochromes b5, NADPH-cytochrome P450 reductases, and family 1 glycosyltransferases[J]. Phytochemistry, 2009, 70(17): 1940-1947. |
| 43 | Willits M G, Giovanni Maité, Prata R T N, et al. Bio-fermentation of modified flavonoids: an example of in vivo diversification of secondary metabolites[J]. Phytochemistry, 2004, 65(1): 31-41. |
| 44 | Lim E K, Ashford D A, Hou B, et al. Arabidopsis glycosyltransferases as biocatalysts in fermentation for regioselective synthesis of diverse quercetin glucosides[J]. Biotechnol. Bioeng., 2004, 87(5): 623-631. |
| 45 | Xia T, Eiteman M A. Quercetin glucoside production by engineered Escherichia coli[J]. Appl. Biochem. Biotechnol., 2017, 182(4): 1358-1370. |
| 46 | Kim B G, Kim H J, Ahn J H. Production of bioactive flavonol rhamnosides by expression of plant genes in Escherichia coli[J]. J. Agric. Food Chem., 2012, 60(44): 11143-11148. |
| 47 | Bruyn F D, Brempt M V, Maertens J, et al. Metabolic engineering of Escherichia coli into a versatile glycosylation platform: production of bio-active quercetin glycosides[J]. Microb. Cell Fact., 2015, 14(1): 138. |
| 48 | Pei J, Chen A, Zhao L, et al. One-pot synthesis of hyperoside by a three-enzyme cascade using a UDP-galactose regeneration system[J]. J. Agric. Food Chem., 2017, 65(29): 6042-6048. |
| 49 | Pandey R P, Malla S, Simkhada D, et al. Production of 3-O-xylosyl quercetin in Escherichia coli[J]. Appl. Microbiol. Biotechnol., 2013, 97(5): 1889-1901. |
| 50 | Han S H, Kim B G, Yoon J A, et al. Synthesis of flavonoid O-pentosides by Escherichia coli through engineering of nucleotide sugar pathways and glycosyltransferase[J]. Appl. Environ. Microbiol., 2014, 80(9): 2754-2762. |
| 51 | Kim S Y, Lee H R, Park K S, et al. Metabolic engineering of Escherichia coli for the biosynthesis of flavonoid-O-glucuronides and flavonoid-O-galactoside[J]. Appl. Microbiol. Biotechnol., 2015, 99(5): 2233-2242. |
| 52 | Kim B G, Sung S H, Ahn J H. Biological synthesis of quercetin 3-O-N-acetylglucosamine conjugate using engineered Escherichia coli expressing UGT78D2[J]. Appl. Microbiol. Biotechnol., 2012, 93(6): 2447-2453. |
| 53 | Roepke J, Bozzo G G. Biocatalytic synthesis of quercetin 3-O-glucoside-7-O-rhamnoside by metabolic engineering of Escherichia coli[J]. ChemBioChem, 2013, 14(18): 2418-2422. |
| 54 | An D G, Yang S M, Kim B G, et al. Biosynthesis of two quercetin O-diglycosides in Escherichia coli[J]. J. Ind. Microbiol. Biotechnol., 2016, 43(6): 841-849. |
| 55 | Choi G S, Kim H J, Kim E J, et al. Stepwise synthesis of quercetin bisglycosides using engineered Escherichia coli[J]. J. Microbiol. Biotechnol., 2018, 28(11): 1859-1864. |
| 56 | Osmani S A, Bak S, Møller B L. Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modeling[J]. Phytochemistry, 2009, 70: 325-347. |
| 57 | Kim M J, Kim B G, Ahn J H. Biosynthesis of bioactive O-methylated flavonoids in Escherichia coli[J]. Appl. Microbiol. Biotechnol., 2013, 97: 7195-7204. |
| 58 | Noguchi A, Horikawa M, Fukui Y, et al. Local differentiation of sugar donor specificity of flavonoid glycosyltransferase in Lamiales[J]. Plant Cell, 2009, 21(5): 1556-1572. |
| 59 | Osmani S A, Bak S, Imberty A, et al. Catalytic key amino acids and UDP-sugar donor specificity of a plant glucuronosyltransferase, UGT94B1: molecular modeling substantiated by site-specifific mutagenesis and biochemical analyses[J]. Plant Physiol., 2008, 148(3): 1295-1308. |
| 60 | Kim B G, Jung N R, Joe E J, et al. Bacterial synthesis of a flavonoid deoxyaminosugar conjugate in Escherichia coli expressing a glycosyltransferase of Arabidopsis thaliana[J]. ChemBioChem, 2010, 11: 2389-2392. |
| 61 | Crh R, Whitfield C. Lipopolysaccharide endotoxins[J]. Annu. Rev. Biochem., 2002, 71: 635-700. |
| 62 | Steiner K, Schwab H. Recent advances in rational approaches for enzyme engineering[J]. Comput. Struct. Biotec., 2012, 2(3): 1-12. |
| 63 | Shao H. Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula[J]. Plant Cell, 2005, 17(11): 3141-3154. |
| 64 | He X Z, Wang X, Dixon R A. Mutational analysis of the Medicago glycosyltransferase UGT71G1 reveals residues that control regioselectivity for (iso)flavonoid glycosylation[J]. J. Biol. Chem., 2006, 281(45): 34441-34447. |
| 65 | Cartwright A M, Lim E K, Kleanthous C, et al. A kinetic analysis of regiospecific glucosylation by two glycosyltransferases of Arabidopsis thaliana: domain swapping to introduce new activities[J]. J. Biol. Chem., 2008, 283(23): 15724-15731. |
| 66 | Chen X. Fermenting next generation glycosylated therapeutics[J]. ACS Chem. Biol., 2011, 6(1): 14-17. |
| 67 | Williams G J, Yang J, Zhang C, et al. Recombinant E. coli prototype strains for in vivo glycorandomization[J]. ACS Chem. Biol., 2011, 6(1): 95-100. |
| 68 | Simkhada D, Kim E M, Lee H C, et al. Metabolic engineering of Escherichia coli for the biological synthesis of 7-O-xylosyl naringenin[J]. Mol. Cells., 2009, 28(4): 397-401. |
| 69 | Malla S, Pandey R P, Kim B G, et al. Regiospecific modifications of naringenin for astragalin production in Escherichia coli[J]. Biotechnol. Bioeng., 2013, 110(9): 2525-2535. |
| 70 | de Bruyn F, de Paepe B, Maertens J, et al. Development of an in vivo glucosylation platform by coupling production to growth: production of phenolic glucosides by a glycosyltransferase of Vitis vinifera[J]. Biotechnol. Bioeng., 2015, 112(8): 1594-1603. |
| 71 | Masada S, Kawase Y, Nagatoshi M, et al. An efficient chemoenzymatic production of small molecule glucosides with in situ UDP-glucose recycling[J]. FEBS Lett., 2007, 581: 2562-2566. |
| 72 | Chen X, Fang J, Zhang J, et al. Sugar nucleotides regeneration beads (superbeads): a versatile tool for the practical synthesis of oligosaccharides[J]. J. Am. Chem. Soc., 2001, 123: 2081-2082. |
| 73 | Lee D C, Cottrill M A, Forsberg C W, et al. Functional insights revealed by the crystal structures of Escherichia coli glucose-1-phosphatase[J]. J. Biol. Chem., 2003, 278(33): 31412-31418. |
| 74 | Oka T, Nemoto T, Jigami Y. Functional analysis of Arabidopsis thaliana RHM2/MUM4, a multidomain protein involved in UDP-D-glucose to UDP-L-rhamnose conversion[J]. J. Biol. Chem., 2007, 282(8): 5389-5403. |
| 75 | Xie Z X, Li B Z, Mitchell L A, et al. “Perfect” designer chromosome V and behavior of a ring derivative [J]. Science, 2017, 355(6329): eaaf4704. |
| 76 | Wu Y, Li B Z, Zhao M, et al. Bug mapping and fitness testing of chemically synthesized chromosome X [J]. Science, 2017, 355(6329): eaaf4706. |
| [1] | 孟令玎, 崇汝青, 孙菲雪, 孟子晖, 刘文芳. 改性聚乙烯膜和氧化硅固定化碳酸酐酶[J]. 化工学报, 2023, 74(8): 3472-3484. |
| [2] | 陈雅鑫, 袁航, 刘冠章, 毛磊, 杨纯, 张瑞芳, 张光亚. 蛋白质纳米笼介导的酶自固定化研究进展[J]. 化工学报, 2023, 74(7): 2773-2782. |
| [3] | 汤晓玲, 王嘉瑞, 朱玄烨, 郑仁朝. 基于Pickering乳液的卤醇脱卤酶催化合成手性环氧氯丙烷[J]. 化工学报, 2023, 74(7): 2926-2934. |
| [4] | 毛磊, 刘冠章, 袁航, 张光亚. 可捕集CO2的纳米碳酸酐酶粒子的高效制备及性能研究[J]. 化工学报, 2023, 74(6): 2589-2598. |
| [5] | 张兰河, 赖青燚, 王铁铮, 关潇卓, 张明爽, 程欣, 徐小惠, 贾艳萍. H2O2对SBR脱氮效率和污泥性能的影响[J]. 化工学报, 2023, 74(5): 2186-2196. |
| [6] | 赵春雷, 郭亮, 高聪, 宋伟, 吴静, 刘佳, 刘立明, 陈修来. 代谢工程改造大肠杆菌生产软骨素[J]. 化工学报, 2023, 74(5): 2111-2122. |
| [7] | 贾露凡, 王艺颖, 董钰漫, 李沁园, 谢鑫, 苑昊, 孟涛. 微流控双水相贴壁液滴流动强化酶促反应研究[J]. 化工学报, 2023, 74(3): 1239-1246. |
| [8] | 刘瑞琪, 周栖桐, 张悦, 贺莹, 高静, 马丽. 基于金纳米颗粒修饰二氧化硅纳米花的生物传感器构建及应用[J]. 化工学报, 2023, 74(3): 1247-1259. |
| [9] | 苏伟怡, 丁佳慧, 李春利, 王洪海, 姜艳军. 酶促反应结晶研究进展[J]. 化工学报, 2023, 74(2): 617-629. |
| [10] | 毕浩然, 张洋, 王凯, 徐晨晨, 霍奕影, 陈必强, 谭天伟. 微生物制造绿色化学品研究进展[J]. 化工学报, 2023, 74(1): 1-13. |
| [11] | 胡阳, 孙彦. 酶分子的自驱动及其介导的微纳马达[J]. 化工学报, 2023, 74(1): 116-132. |
| [12] | 谭卓涛, 齐思雨, 许梦蛟, 戴杰, 朱晨杰, 应汉杰. 辅酶自循环的氧化还原级联体系在生物催化过程中的应用:机遇与挑战[J]. 化工学报, 2023, 74(1): 45-59. |
| [13] | 刘雪, 张莉娟, 赵广荣. 大肠杆菌偏利共培养系统合成大豆苷元[J]. 化工学报, 2022, 73(9): 4015-4024. |
| [14] | 安绍杰, 许洪峰, 李思, 许远航, 李佳锡. 利用分子机器的组装与分解构建pH敏感性谷胱甘肽过氧化物人工酶[J]. 化工学报, 2022, 73(8): 3669-3678. |
| [15] | 张昕哲, 孙文涛, 吕波, 李春. 植物天然产物氧化与微生物制造[J]. 化工学报, 2022, 73(7): 2790-2805. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备 11010102001995号
京公网安备 11010102001995号