化工学报 ›› 2021, Vol. 72 ›› Issue (7): 3757-3767.DOI: 10.11949/0438-1157.20201841
收稿日期:2020-12-16
修回日期:2021-01-27
出版日期:2021-07-05
发布日期:2021-07-05
通讯作者:
王世珍
作者简介:段凌暄(1998—),女,硕士研究生,基金资助:
DUAN Lingxuan( ),YAO Guangxiao,JIANG Liang,WANG Shizhen(
),YAO Guangxiao,JIANG Liang,WANG Shizhen( )
)
Received:2020-12-16
Revised:2021-01-27
Online:2021-07-05
Published:2021-07-05
Contact:
WANG Shizhen
摘要:
可在非水相体系高效催化不对称还原反应制备手性化合物的氧化还原酶具有重要科学意义与工业应用前景。基于基因挖掘技术,获得了17个耐盐氨基酸脱氢酶的基因,并分析了其进化同源性和蛋白质稳定性热力学参数。选取来源于Natranaerobius thermophilus的苯丙氨酸脱氢酶(PheDH),进行了基因合成和表达、分离和纯化,获得了耐盐氨基酸脱氢酶,并检测了其有机溶剂耐受性。结果表明,对于催化L-苯丙氨酸的氧化脱氨体系,反应的最适温度为60℃,最适pH为 12。在含有30%的二甲亚砜反应体系中,催化活性是水相体系的1.2倍。而对于催化还原胺化制备L-高苯丙氨酸的体系,最适温度为70℃,最适pH为8.5。在含30%的甲基叔丁基醚和二甲亚砜反应体系中,催化活性分别是原始活性的101.3%和99.2%。研究表明,该耐盐酶具有较好的耐热、耐有机溶剂等抗逆性能。
中图分类号:
段凌暄, 姚光晓, 江亮, 王世珍. 耐有机溶剂氨基酸脱氢酶基因挖掘与非天然氨基酸的非水相合成[J]. 化工学报, 2021, 72(7): 3757-3767.
DUAN Lingxuan, YAO Guangxiao, JIANG Liang, WANG Shizhen. Genome mining of organic solvent tolerant amino acid dehydrogenase for biosynthesis of unnatural amino acids in non-aqueous system[J]. CIESC Journal, 2021, 72(7): 3757-3767.
| 分类 | 菌株名称 | 分类 | 菌株名称 |
|---|---|---|---|
| 嗜热 | Geobacillus kaustophilus | 嗜盐 | Halobacterium salinarum |
| Natranaerobius thermophilus | Haloferax volcanii | ||
| Methanococcus jannaschii | Haloferax lucentense | ||
| Thermotoga maritima | Haloarcula japonica | ||
| Geobacillus stearothermophilus 10 | Halogranum rubrum | ||
| Sulfobacillus thermosulfidooxidans | Halogeometyicum borinquense | ||
| 嗜热嗜碱 | Anaerobranca gottschalkii | 嗜盐嗜碱 | Euhalothece natronophila |
| Halothermothrix orenii | Ferroplasma acidiphilum |
表1 筛选获得的来源于极端微生物的氨基酸脱氢酶
Table 1 Screening of amino acid dehydrogenases from extremophiles
| 分类 | 菌株名称 | 分类 | 菌株名称 |
|---|---|---|---|
| 嗜热 | Geobacillus kaustophilus | 嗜盐 | Halobacterium salinarum |
| Natranaerobius thermophilus | Haloferax volcanii | ||
| Methanococcus jannaschii | Haloferax lucentense | ||
| Thermotoga maritima | Haloarcula japonica | ||
| Geobacillus stearothermophilus 10 | Halogranum rubrum | ||
| Sulfobacillus thermosulfidooxidans | Halogeometyicum borinquense | ||
| 嗜热嗜碱 | Anaerobranca gottschalkii | 嗜盐嗜碱 | Euhalothece natronophila |
| Halothermothrix orenii | Ferroplasma acidiphilum |
| Number | Average value | ΔHm/(kcal/mol) | ΔCp/(kcal/(mol·K)) | Tm/℃ | ΔGr/(kcal/mol) |
|---|---|---|---|---|---|
| 1 | KJE28589.1 | -144.7 | -4.02 | 67.1 | -7 |
| 2 | WP-010871127.1 | -127.1 | -3.65 | 65 | -6 |
| 3 | WP-091347685.1 | -138 | -3.6 | 71.2 | -6.8 |
| 4 | WP-004590695.1 | -154 | -4.36 | 65.9 | -7.4 |
| 5 | ALA71326.1 | -118.5 | -3.49 | 63.4 | -5.6 |
| 6 | PSR36482.1 | -128 | -3.42 | 69.6 | -6.3 |
| 7 | WP-012635747.1 | -95.9 | -2.71 | 66 | -4.6 |
| 8 | NT2349 | -113.3 | -3.34 | 63.5 | -5.3 |
表2 不同极端菌株来源的氨基酸脱氢酶的热力学参数比较
Table 2 Comparison of thermodynamic parameters of amino acid dehydrogenases from extremophiles
| Number | Average value | ΔHm/(kcal/mol) | ΔCp/(kcal/(mol·K)) | Tm/℃ | ΔGr/(kcal/mol) |
|---|---|---|---|---|---|
| 1 | KJE28589.1 | -144.7 | -4.02 | 67.1 | -7 |
| 2 | WP-010871127.1 | -127.1 | -3.65 | 65 | -6 |
| 3 | WP-091347685.1 | -138 | -3.6 | 71.2 | -6.8 |
| 4 | WP-004590695.1 | -154 | -4.36 | 65.9 | -7.4 |
| 5 | ALA71326.1 | -118.5 | -3.49 | 63.4 | -5.6 |
| 6 | PSR36482.1 | -128 | -3.42 | 69.6 | -6.3 |
| 7 | WP-012635747.1 | -95.9 | -2.71 | 66 | -4.6 |
| 8 | NT2349 | -113.3 | -3.34 | 63.5 | -5.3 |

图2 NT2349的结构和表面电荷分布和带电性能图(a) NT2349的三维结构; (b) 酸性和碱性氨基酸残基分布(红色代表酸性氨基酸,蓝色代表碱性氨基酸); (c) NT2349的表面电荷分布(红色代表负电荷,蓝色代表正电荷)
Fig.2 Structure of NT2349 and surface charged residues distribution(a) NT2349 three-dimensional structure; (b) acidic and basic amino acid residue distribution (red represents acidic amino acids, blue represents basic amino acids); (c) the surface charge distribution of NT2349 (red represents negative charge, blue represents positive charge)
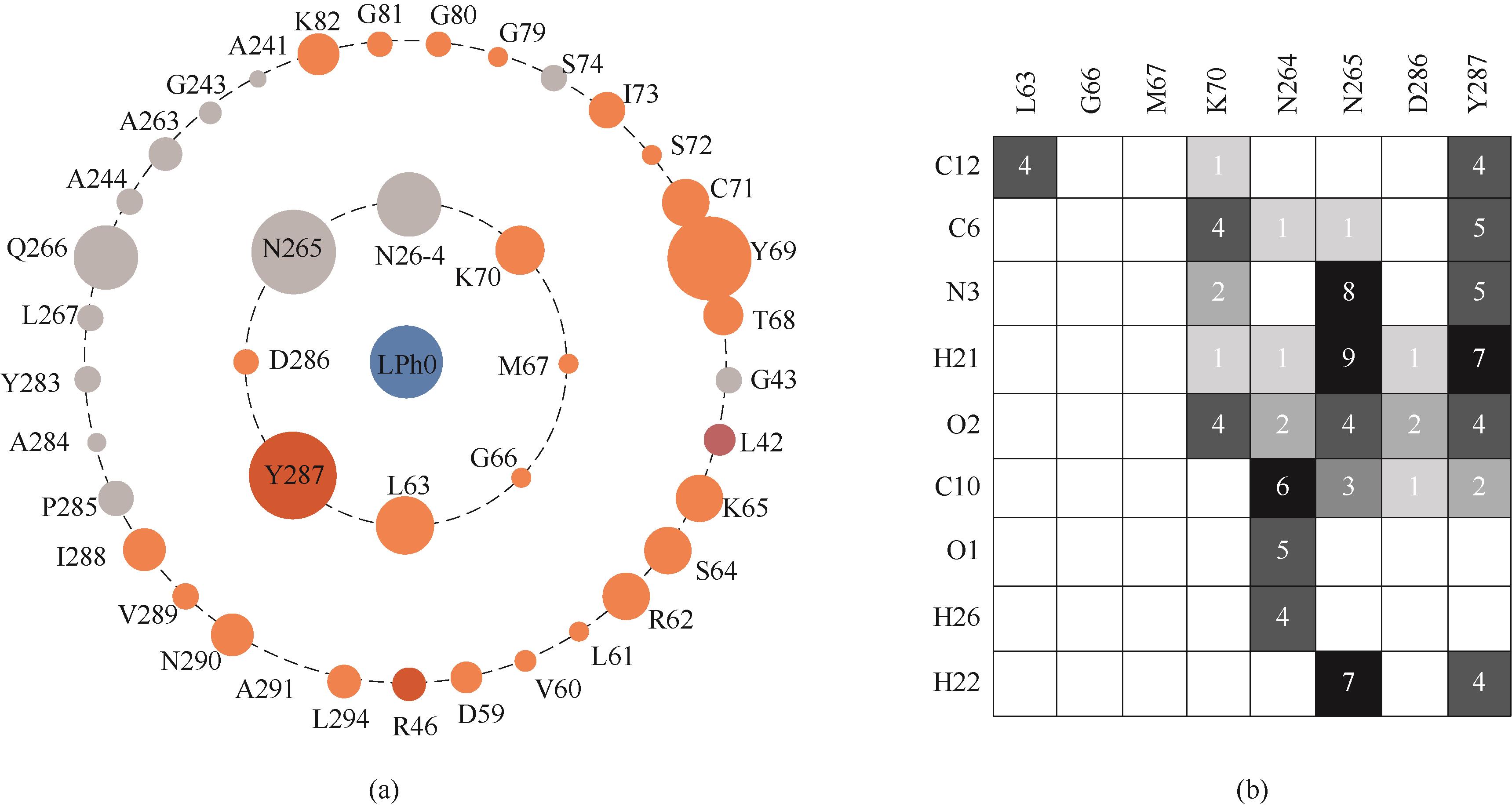
图3 NT2349与底物的二级结构相互作用图(a) NT2349的氨基酸残基与苯丙氨酸相互作用图;(b) NT2349活性位点与苯丙氨酸相互作用的Heatmap图
Fig.3 Interactions of secondary structure of NT2349 with L-phenylalanine(a) key residues interact with L-phenylalanine; (b) interaction Heatmap of active site with L-phenylalanine
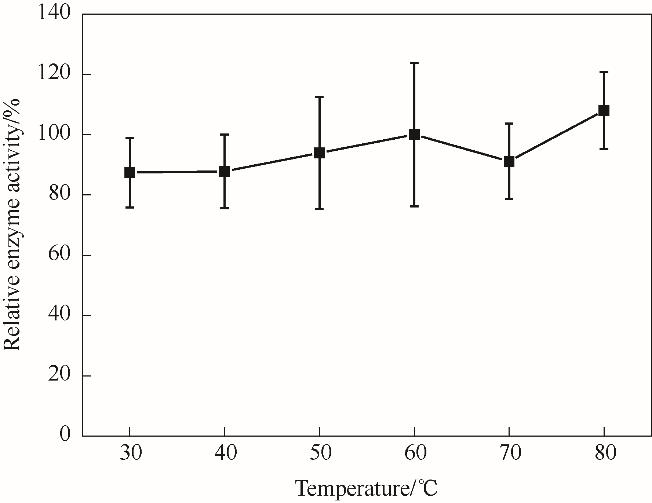
图6 温度对NT2349的氧化脱氨活性影响Reaction conditions: L-Phe, 2 mmol/L; glycine-NaOH buffer, 160 mmol/L (pH 10); NAD+, 0.2 mmol/L
Fig.6 Effect of temperature on oxidative deamination activity of NT2349
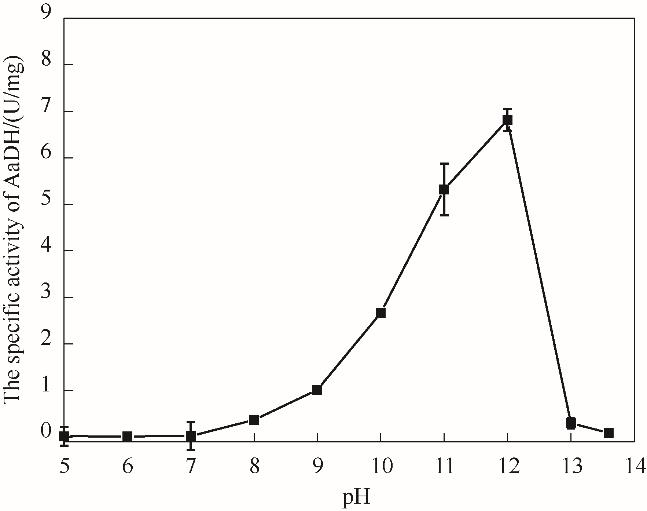
图7 pH对NT2349的氧化脱氨活性影响Reaction conditions: L-Phe, 2 mmol/L; 0.2 mol/L potassium phosphate buffer, pH 5—6; 0.2 mol/L Tris-HCl buffer, pH 7—8; 0.2mol/L glycine-NaOH buffer, pH 9—13.6 ;NAD+, 0.2 mmol/L
Fig.7 The effect of pH on oxidative deamination activity of NT2349
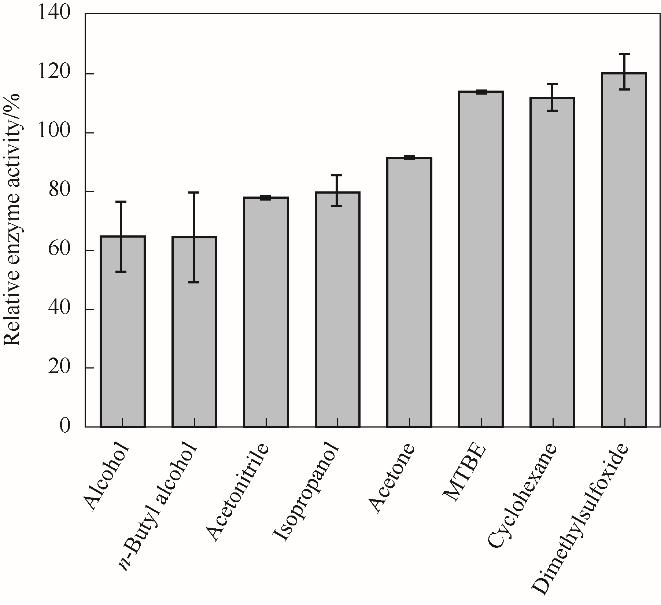
图8 有机溶剂对NT2349的氧化脱氨反应酶活影响Reaction conditions: L-Phe, 2 mmol/L; glycine-NaOH buffer, 160 mmol/L (pH 10); NAD+, 0.2 mmol/L
Fig.8 Effect of organic solvent on oxidative deamination activity of NT2349
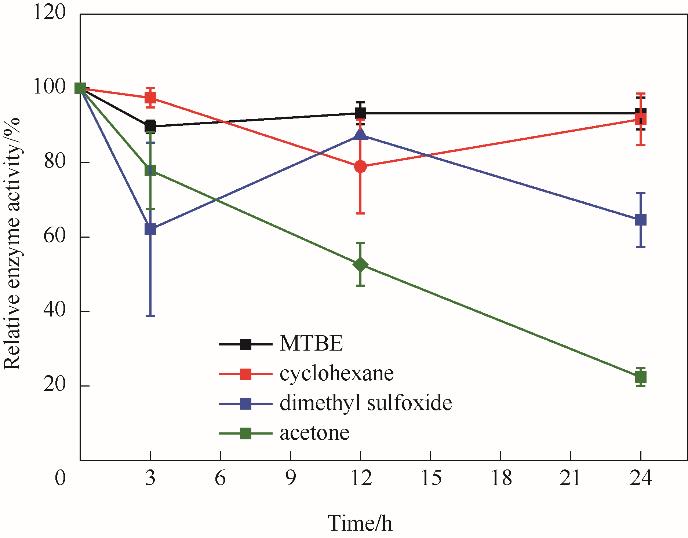
图9 有机溶剂体系中酶催化氧化脱氨的酶活稳定性Reaction conditions: L-Phe, 2 mmol/L; glycine-NaOH buffer, 160 mmol/L (pH 10); NAD+,0.2 mmol/L
Fig.9 Enzyme activity stability of oxidative deamination of NT2349 in organic solvents
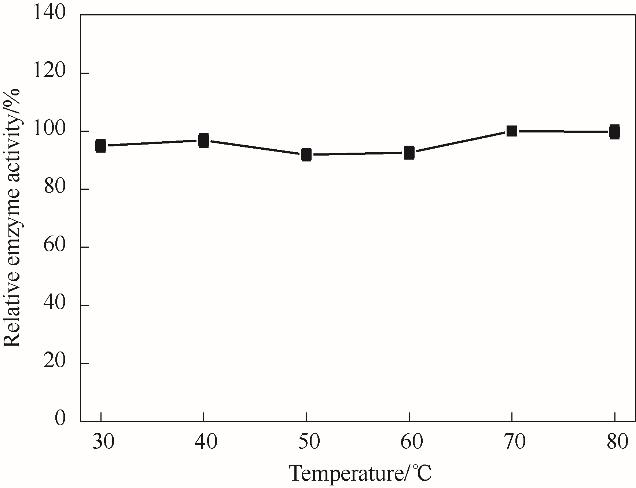
图11 温度对NT2349的还原胺化活性影响Reaction conditions: EOPB, 2 mmol/L; NH4Cl-NH3·H2O buffer, 160 mmol/L (pH 8.5); NADH 0.2 mmol/L
Fig.11 The effect of temperature on the reductive amination activity of NT2349
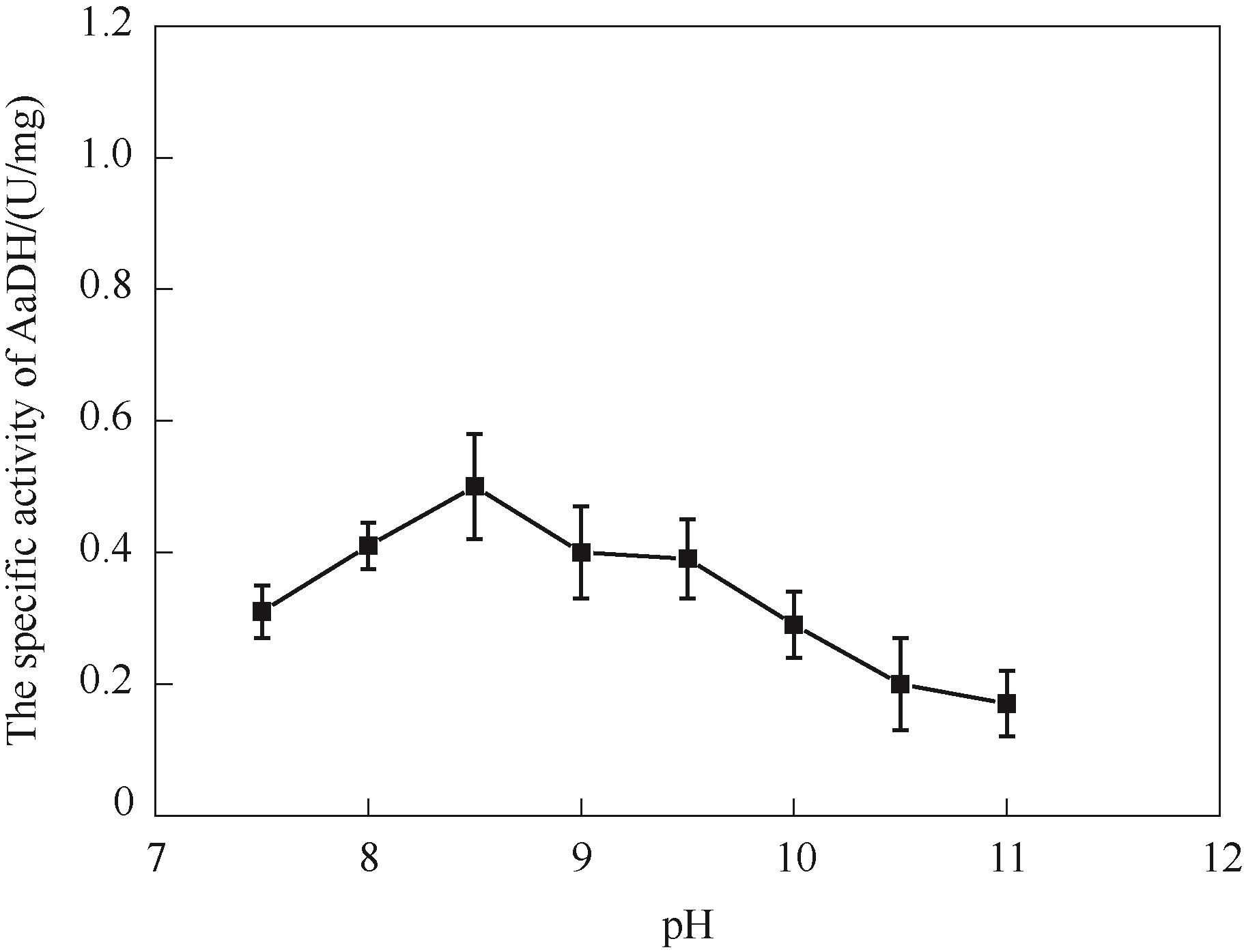
图12 pH对NT2349的还原胺化活性影响Reaction conditions: EOPB, 2 mmol/L; NH4Cl-NH3·H2O buffer, 160 mmol/L (pH 7.5—11); NADH 0.2 mmol/L
Fig.12 The effect of pH on the reductive amination activity of NT2349

图13 有机溶剂对NT2349的还原胺化反应酶活影响Reaction conditions: EOPB, 2 mmol/L; NH4Cl-NH3·H2O buffer, 160 mmol/L (pH 8.5); NADH 0.2 mmol/L
Fig.13 Effect of organic solvent on the activity of reductive ammoniation reactivity
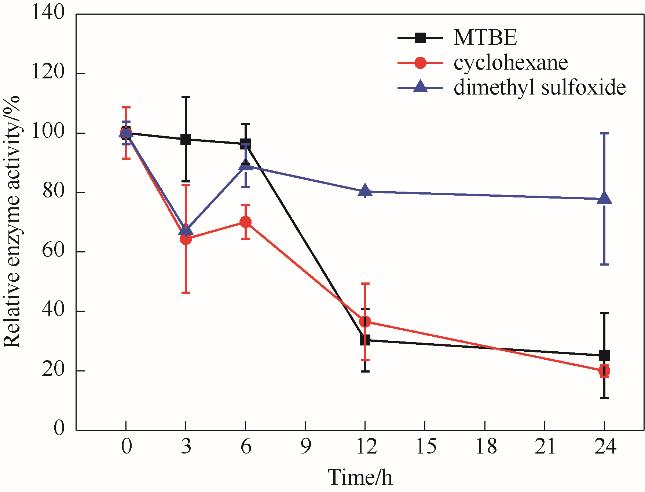
图14 有机溶剂中NT2349催化还原胺化的酶活稳定性Reaction conditions: EOPB, 2 mmol/L; NH4Cl-NH3·H2O buffer, 160 mmol/L (pH 8.5); NADH, 0.2 mmol/L
Fig.14 Enzyme activity stability of NT2349 reductive amination in organic solvents
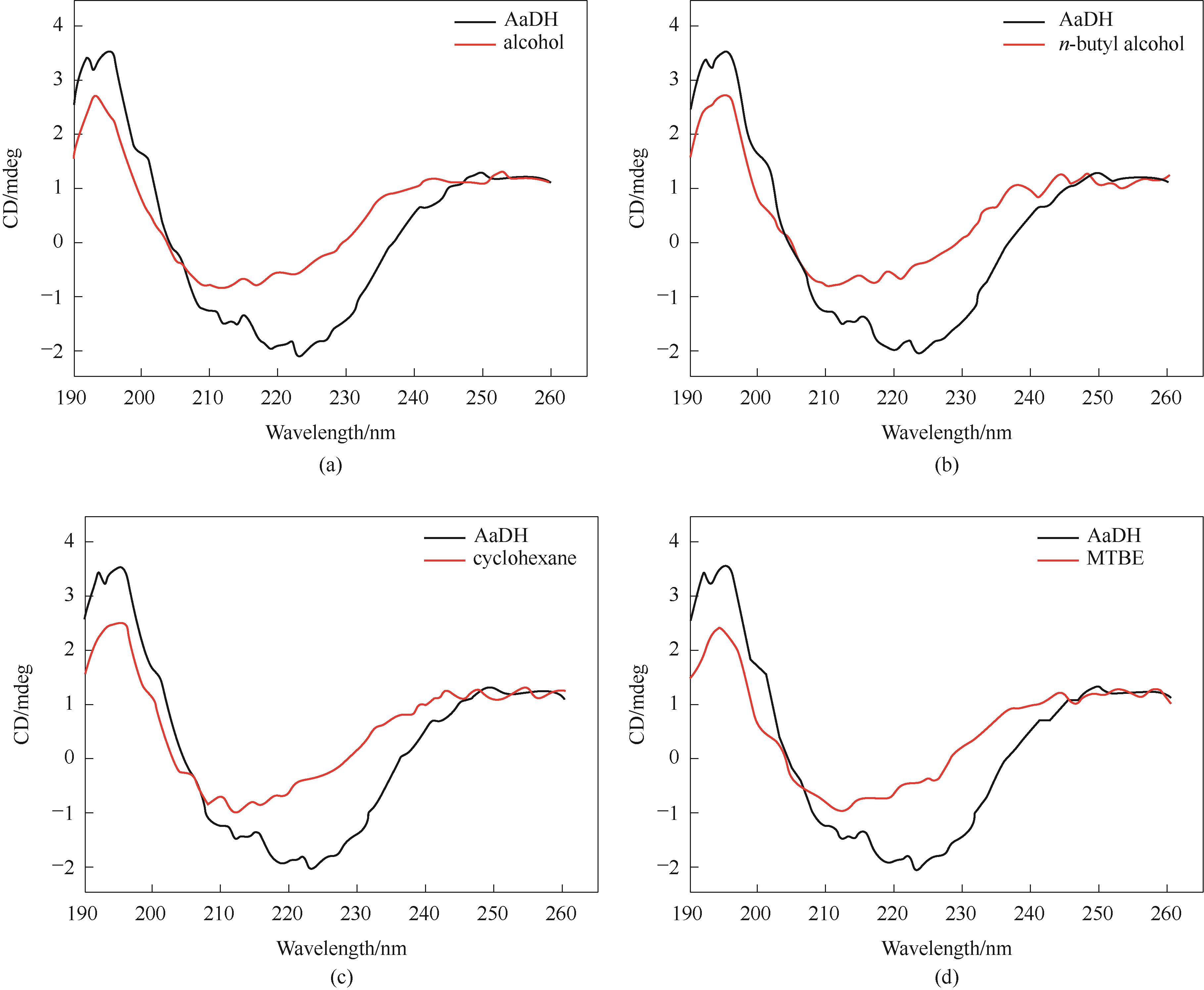
图15 NT2349在30%有机溶剂下的圆二色谱图(a)乙醇; (b)正丁醇; (c)环己烷; (d)甲基叔丁基醚
Fig.15 Circular dichroism spectra of NT2349 in 30% organic solvents(a) alcohol; (b) n-butyl alcohol; (c) cyclohexane; (d) MTBE
| 1 | Liszka M J, Clark M E, Schneider E, et al. Nature versus nurture: developing enzymes that function under extreme conditions[J]. Annual Review of Chemical and Biomolecular Engineering, 2012, 3: 77-102. |
| 2 | Chen F F, Zheng G W, Liu L, et al. Reshaping the active pocket of amine dehydrogenases for asymmetric synthesis of bulky aliphatic amines[J]. ACS Catalysis, 2018, 8(3): 2622-2628. |
| 3 | Dhake K P, Thakare D D, Bhanage B M. Lipase: a potential biocatalyst for the synthesis of valuable flavour and fragrance ester compounds[J]. Flavour and Fragrance Journal, 2013, 28(2): 71-83. |
| 4 | DasSarma S, DasSarma P. Halophiles and their enzymes: negativity put to good use[J]. Current Opinion in Microbiology, 2015, 25: 120-126. |
| 5 | Chen S, Engel P. An engineered mutant, L307V of phenylalanine dehydrogenase from Bacillus sphaericus: high activity and stability in organic-aqueous solvent mixtures and utility for synthesis of non-natural L-amino acids [J]. Journal of Biotechnology,2007, 131(2): 116. |
| 6 | Munawar N, Engel P C. Overexpression in a non-native halophilic host and biotechnological potential of NAD+-dependent glutamate dehydrogenase from Halobacterium salinarum strain NRC-36014[J]. Extremophiles, 2012, 16(3): 463-476. |
| 7 | Alsafadi D, Paradisi F. Effect of organic solvents on the activity and stability of halophilic alcohol dehydrogenase (ADH2) from Haloferax volcanii[J]. Extremophiles, 2013, 17(1): 115-122. |
| 8 | Munawar N, Engel P C. Prospects for robust biocatalysis: engineering of novel specificity in a halophilic amino acid dehydrogenase[J]. Extremophiles, 2013, 17(1): 43-51. |
| 9 | Timpson L M, Alsafadi D, Mac Donnchadha C, et al. Characterization of alcohol dehydrogenase (ADH12) from Haloarcula marismortui, an extreme halophile from the Dead Sea[J]. Extremophiles, 2012, 16(1): 57-66. |
| 10 | Hartmann E M, Durighello E, Pible O, et al. Proteomics meets blue biotechnology: a wealth of novelties and opportunities[J]. Marine Genomics, 2014, 17: 35-42. |
| 11 | Nguyen V, Wilson C, Hoemberger M, et al. Evolutionary drivers of thermoadaptation in enzyme catalysis[J]. Science, 2017, 355(6322): 289-294. |
| 12 | Broom A, Trainor K, MacKenzie D W, et al. Using natural sequences and modularity to design common and novel protein topologies[J]. Current Opinion in Structural Biology, 2016, 38: 26-36. |
| 13 | Wang H Y, Qu G, Li J K, et al. Data mining of amine dehydrogenases for the synthesis of enantiopure amino alcohols[J]. Catalysis Science & Technology, 2020, 10(17): 5945-5952. |
| 14 | Dalmaso G Z, Ferreira D, Vermelho A B. Marine extremophiles: a source of hydrolases for biotechnological applications[J]. Marine Drugs, 2015, 13(4): 1925-1965. |
| 15 | Trincone A. Biocatalytic processes using marine biocatalysts: ten cases in point[J]. Current Organic Chemistry, 2013, 17(10): 1058-1066. |
| 16 | Mutti F G, Knaus T, Scrutton N S, et al. Conversion of alcohols to enantiopure amines through dual-enzyme hydrogen-borrowing cascades[J]. Science, 2015, 349(6255): 1525-1529. |
| 17 | Ducrot L, Bennett M, Grogan G, et al. NAD(P)H-dependent enzymes for reductive amination: active site description and carbonyl-containing compound spectrum[J]. Advanced Synthesis & Catalysis, 2021, 363(2): 328-351. |
| 18 | Liu Z N, Lei D W, Qiao B, et al. Integrative biosynthetic gene cluster mining to optimize a metabolic pathway to efficiently produce L-homophenylalanine in Escherichia coli[J]. ACS Synthetic Biology, 2020, 9(11): 2943-2954. |
| 19 | 陈曦, 高秀珍, 朱敦明. 氨基酸脱氢酶的催化机理、分子改造及合成应用[J]. 微生物学报, 2017, 57(8): 1249-1261. |
| Chen X, Gao X Z, Zhu D M. Catalysis, engineering and application of amino acid dehydrogenases[J]. Acta Microbiologica Sinica, 2017, 57(8): 1249-1261. | |
| 20 | Vallenet D, Calteau A, Cruveiller S, et al. MicroScope in 2017: an expanding and evolving integrated resource for community expertise of microbial genomes[J]. Nucleic Acids Research, 2017, 45(D1): D517-D528. |
| 21 | Schaeffer D, Grishin N V. Identification of protein homologs and domain boundaries by iterative sequence alignment [J]. Computational Methods in Protein Evolution, 2019, 1851: 277-286. |
| 22 | Yang J Y, Zhang Y. I-TASSER server: new development for protein structure and function predictions[J]. Nucleic Acids Research, 2015, 43(W1): W174-W181. |
| 23 | Studer G, Rempfer C, Waterhouse A M, et al. QMEANDisCo—distance constraints applied on model quality estimation[J]. Bioinformatics, 2020, 36(6): 1765-1771. |
| 24 | Kikani B A, Singh S P. The stability and thermodynamic parameters of a very thermostable and calcium-independent α-amylase from a newly isolated bacterium, Anoxybacillus beppuensis TSSC-1[J]. Process Biochemistry, 2012, 47(12): 1791-1798. |
| 25 | Lin P, Zhang Y H, Yao G X, et al. Immobilization of formate dehydrogenase on polyethylenimine-grafted graphene oxide with kinetics and stability study[J]. Engineering in Life Sciences, 2020, 20(3/4): 104-111. |
| 26 | 石云云, 李信志, 张桂敏. 微生物嗜盐酶的研究进展[J]. 微生物学报, 2017, 57(8): 1180-1188. |
| Shi Y Y, Li X Z, Zhang G M. Advances in microbial halophilic enzymes[J]. Acta Microbiologica Sinica, 2017, 57(8): 1180-1188. | |
| 27 | Kayikci M, Venkatakrishnan A J, Scott-Brown J, et al. Visualization and analysis of non-covalent contacts using the Protein Contacts Atlas[J]. Nature Structural & Molecular Biology, 2018, 25(2): 185-194. |
| 28 | Xu L S, Wang Z Y, Mao P T, et al. Enzymatic synthesis of S-phenyl-L-cysteine from keratin hydrolysis industries wastewater with tryptophan synthase[J]. Bioresource Technology, 2013, 133: 635-637. |
| 29 | Dumorné K, Córdova D C, Astorga-Eló M, et al. Extremozymes: a potential source for industrial applications[J]. Journal of Microbiology and Biotechnology, 2017, 27(4): 649-659. |
| 30 | Raddadi N, Cherif A, Daffonchio D, et al. Biotechnological applications of extremophiles, extremozymes and extremolytes[J]. Applied Microbiology and Biotechnology, 2015, 99(19): 7907-7913. |
| 31 | Jin M, Gai Y, Guo X, et al. Properties and applications of extremozymes from deep-sea extremophilic microorganisms: a mini review[J]. Marine Drugs, 2019, 17(12): 656. |
| [1] | 程业品, 胡达清, 徐奕莎, 刘华彦, 卢晗锋, 崔国凯. 离子液体基低共熔溶剂在转化CO2中的应用[J]. 化工学报, 2023, 74(9): 3640-3653. |
| [2] | 孟令玎, 崇汝青, 孙菲雪, 孟子晖, 刘文芳. 改性聚乙烯膜和氧化硅固定化碳酸酐酶[J]. 化工学报, 2023, 74(8): 3472-3484. |
| [3] | 陈雅鑫, 袁航, 刘冠章, 毛磊, 杨纯, 张瑞芳, 张光亚. 蛋白质纳米笼介导的酶自固定化研究进展[J]. 化工学报, 2023, 74(7): 2773-2782. |
| [4] | 汤晓玲, 王嘉瑞, 朱玄烨, 郑仁朝. 基于Pickering乳液的卤醇脱卤酶催化合成手性环氧氯丙烷[J]. 化工学报, 2023, 74(7): 2926-2934. |
| [5] | 毛磊, 刘冠章, 袁航, 张光亚. 可捕集CO2的纳米碳酸酐酶粒子的高效制备及性能研究[J]. 化工学报, 2023, 74(6): 2589-2598. |
| [6] | 张兰河, 赖青燚, 王铁铮, 关潇卓, 张明爽, 程欣, 徐小惠, 贾艳萍. H2O2对SBR脱氮效率和污泥性能的影响[J]. 化工学报, 2023, 74(5): 2186-2196. |
| [7] | 刘瑞琪, 周栖桐, 张悦, 贺莹, 高静, 马丽. 基于金纳米颗粒修饰二氧化硅纳米花的生物传感器构建及应用[J]. 化工学报, 2023, 74(3): 1247-1259. |
| [8] | 贾露凡, 王艺颖, 董钰漫, 李沁园, 谢鑫, 苑昊, 孟涛. 微流控双水相贴壁液滴流动强化酶促反应研究[J]. 化工学报, 2023, 74(3): 1239-1246. |
| [9] | 苏伟怡, 丁佳慧, 李春利, 王洪海, 姜艳军. 酶促反应结晶研究进展[J]. 化工学报, 2023, 74(2): 617-629. |
| [10] | 刘昕, 戈钧, 李春. 光驱动微生物杂合系统提高生物制造水平[J]. 化工学报, 2023, 74(1): 330-341. |
| [11] | 胡阳, 孙彦. 酶分子的自驱动及其介导的微纳马达[J]. 化工学报, 2023, 74(1): 116-132. |
| [12] | 谭卓涛, 齐思雨, 许梦蛟, 戴杰, 朱晨杰, 应汉杰. 辅酶自循环的氧化还原级联体系在生物催化过程中的应用:机遇与挑战[J]. 化工学报, 2023, 74(1): 45-59. |
| [13] | 安绍杰, 许洪峰, 李思, 许远航, 李佳锡. 利用分子机器的组装与分解构建pH敏感性谷胱甘肽过氧化物人工酶[J]. 化工学报, 2022, 73(8): 3669-3678. |
| [14] | 张劢, 田瑶, 郭之旗, 王叶, 窦广进, 宋浩. 光催化-生物杂合系统设计优化用于燃料和化学品绿色合成[J]. 化工学报, 2022, 73(7): 2774-2789. |
| [15] | 黄丽菁, 黄继娇, 李鹏辉, 刘芷诺, 蒋康杰, 吴文娟. 木质素羟丙基磺甲基化改性及其对纤维素酶水解的影响[J]. 化工学报, 2022, 73(7): 3232-3239. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备 11010102001995号
京公网安备 11010102001995号